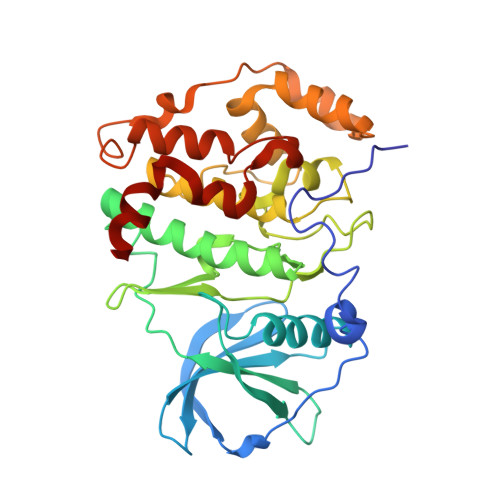
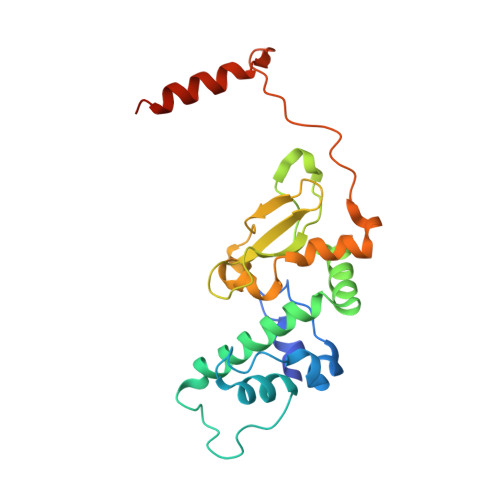
The Protein Kinase CK2(Andante) Holoenzyme Structure Supports Proposed Models of Autoregulation and Trans-Autophosphorylation
Schnitzler, A., Olsen, B.B., Issinger, O.-G., Niefind, K.(2014) J Mol Biology 426: 1871-1882
- PubMed: 24594356
- DOI: https://doi.org/10.1016/j.jmb.2014.02.018
- Primary Citation of Related Structures:
4NH1 - PubMed Abstract:
Eukaryotic protein kinases are typically strictly controlled by second messenger binding, protein/protein interactions, dephosphorylations or similar processes. None of these regulatory mechanisms is known to work for protein kinase CK2 (former name "casein kinase 2"), an acidophilic and constitutively active eukaryotic protein kinase. CK2 predominantly exists as a heterotetrameric holoenzyme composed of two catalytic subunits (CK2α) complexed to a dimer of non-catalytic subunits (CK2β). One model of CK2 regulation was proposed several times independently by theoretical docking of the first CK2 holoenzyme structure. According to this model, the CK2 holoenzyme forms autoinhibitory aggregates correlated with trans-autophosphorylation and driven by the down-regulatory affinity between an acidic loop of CK2β and the positively charged substrate binding region of CK2α from a neighboring CK2 heterotetramer. Circular trimeric aggregates in which one-half of the CK2α chains show the predicted inhibitory proximity between those regions were detected within the crystal packing of the human CK2 holoenzyme. Here, we present further in vitro support of the "regulation-by-aggregation" model by an alternative crystal form in which CK2 tetramers are arranged as approximately linear aggregates coinciding essentially with the early predictions. In this assembly, the substrate binding region of every CK2α chain is blocked by a CK2β acidic loop from a neighboring tetramer. We found these crystals with CK2(Andante) that contains a CK2β variant mutated in a CK2α-contact helix and described to be responsible for a prolonged circadian rhythm in Drosophila. The increased propensity of CK2(Andante) to form aggregates with completely blocked active sites may contribute to this phenotype.
- Institut für Biochemie, Department für Chemie, Universität zu Köln, Otto-Fischer-Straße 12-14, D-50674 Köln, Germany.
Organizational Affiliation: