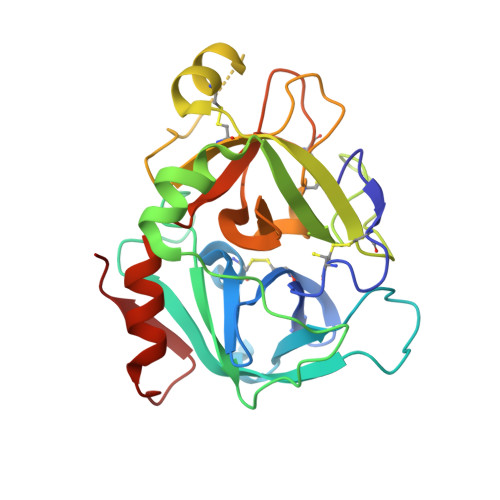
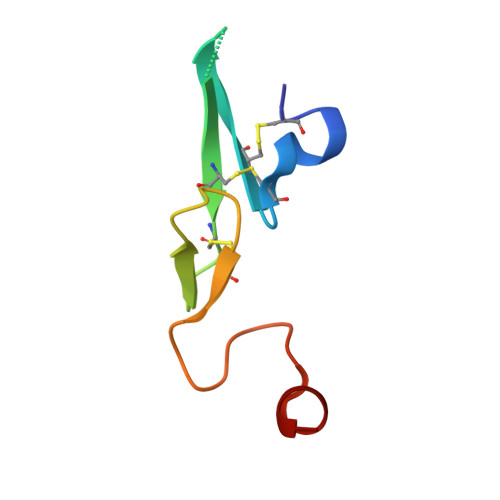
Design and Synthesis of Phenylpyrrolidine Phenylglycinamides As Highly Potent and Selective TF-FVIIa Inhibitors.
Zhang, X., Jiang, W., Jacutin-Porte, S., Glunz, P.W., Zou, Y., Cheng, X., Nirschl, A.H., Wurtz, N.R., Luettgen, J.M., Rendina, A.R., Luo, G., Harper, T.M., Wei, A., Anumula, R., Cheney, D.L., Knabb, R.M., Wong, P.C., Wexler, R.R., Priestley, E.S.(2014) ACS Med Chem Lett 5: 188-192
- PubMed: 24900796
- DOI: https://doi.org/10.1021/ml400453z
- Primary Citation of Related Structures:
4NG9, 4NGA - PubMed Abstract:
Inhibitors of the Tissue Factor/Factor VIIa (TF-FVIIa) complex are promising novel anticoagulants that show excellent efficacy and minimal bleeding in preclinical models. On the basis of a zwitterionic phenylglycine acylsulfonamide 1, a phenylglycine benzylamide 2 was shown to possess improved permeability and oral bioavailability. Optimization of the benzylamide, guided by X-ray crystallography, led to a potent TF-FVIIa inhibitor 18i with promising oral bioavailability, but promiscuous activity in an in vitro safety panel of receptors and enzymes. Introducing an acid on the pyrrolidine ring, guided by molecular modeling, resulted in highly potent, selective, and efficacious TF-FVIIa inhibitors with clean in vitro safety profile. The pyrrolidine acid 20 showed a moderate clearance, low volume of distribution, and a short t 1/2 in dog PK studies.
- Bristol-Myers Squibb R&D , 311 Pennington-Rocky Hill Road, Pennington, New Jersey 08534-2130, United States.
Organizational Affiliation: