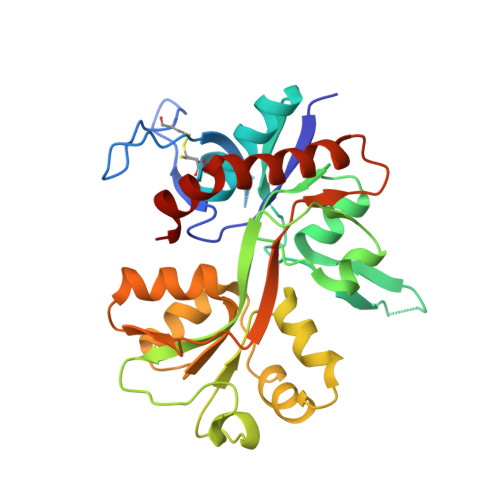
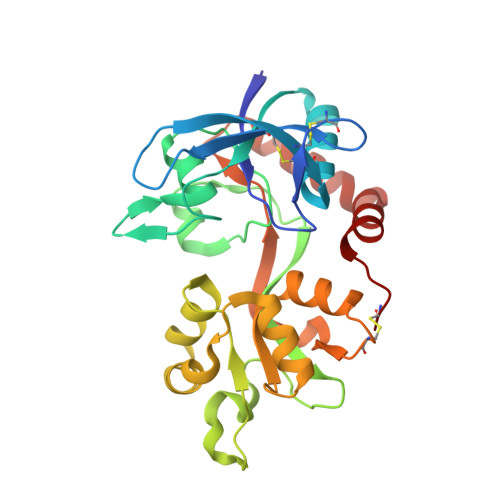
Structural Insights into Competitive Antagonism in NMDA Receptors.
Jespersen, A., Tajima, N., Fernandez-Cuervo, G., Garnier-Amblard, E.C., Furukawa, H.(2014) Neuron 81: 366-378
- PubMed: 24462099
- DOI: https://doi.org/10.1016/j.neuron.2013.11.033
- Primary Citation of Related Structures:
4NF4, 4NF5, 4NF6, 4NF8 - PubMed Abstract:
There has been a great level of enthusiasm to downregulate overactive N-methyl-D-aspartate (NMDA) receptors to protect neurons from excitotoxicity. NMDA receptors play pivotal roles in basic brain development and functions as well as in neurological disorders and diseases. However, mechanistic understanding of antagonism in NMDA receptors is limited due to complete lack of antagonist-bound structures for the L-glutamate-binding GluN2 subunits. Here, we report the crystal structures of GluN1/GluN2A NMDA receptor ligand-binding domain (LBD) heterodimers in complex with GluN1- and GluN2-targeting antagonists. The crystal structures reveal that the antagonists, D-(-)-2-amino-5-phosphonopentanoic acid (D-AP5) and 1-(phenanthrene-2-carbonyl)piperazine-2,3-dicarboxylic acid (PPDA), have discrete binding modes and mechanisms for opening of the bilobed architecture of GluN2A LBD compared to the agonist-bound form. The current study shows distinct ways by which the conformations of NMDA receptor LBDs may be controlled and coupled to receptor inhibition and provides possible strategies to develop therapeutic compounds with higher subtype-specificity.
- Cold Spring Harbor Laboratory, WM Keck Structural Biology Laboratory, 1 Bungtown Road, Cold Spring Harbor, NY 11724, USA.
Organizational Affiliation: