Structural basis for hijacking CBF-b and CUL5 E3 ligase complex by HIV-1 Vif
Guo, Y.Y., Dong, L.Y., Qiu, X.L., Wang, Y.S., Zhang, B.L., Liu, H.N., Yu, Y., Zang, Y., Yang, M.J., Huang, Z.W.(2014) Nature 505: 229-233
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
(2014) Nature 505: 229-233
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cullin-5 | 311 | Homo sapiens | Mutation(s): 0 Gene Names: CUL5, VACM1 | 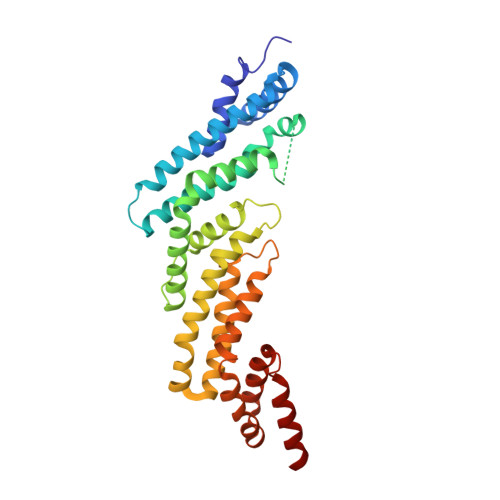 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q93034 (Homo sapiens) Explore Q93034 Go to UniProtKB: Q93034 | |||||
PHAROS: Q93034 GTEx: ENSG00000166266 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q93034 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Transcription elongation factor B polypeptide 1 | 96 | Homo sapiens | Mutation(s): 0 Gene Names: ELOC, TCEB1 | 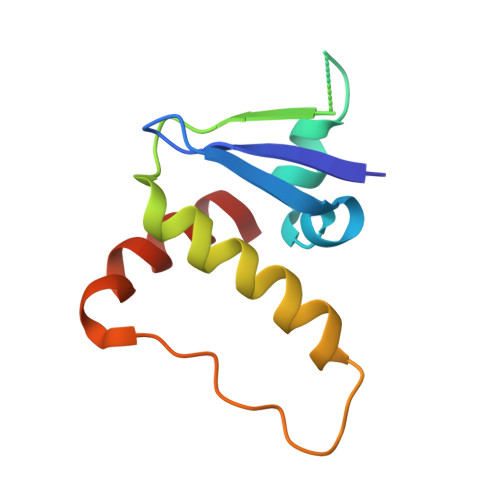 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q15369 (Homo sapiens) Explore Q15369 Go to UniProtKB: Q15369 | |||||
PHAROS: Q15369 GTEx: ENSG00000154582 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q15369 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Transcription elongation factor B polypeptide 2 | 102 | Homo sapiens | Mutation(s): 0 Gene Names: ELOB, TCEB2 | 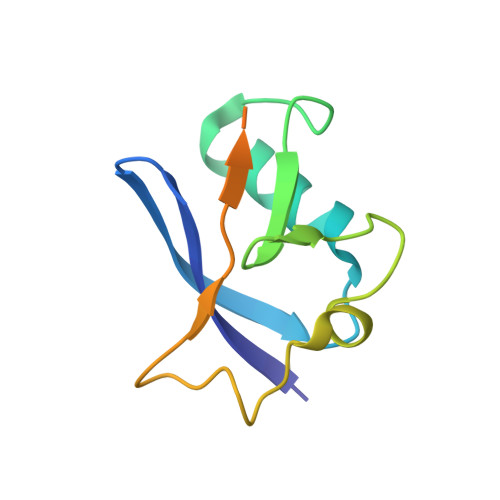 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q15370 (Homo sapiens) Explore Q15370 Go to UniProtKB: Q15370 | |||||
PHAROS: Q15370 GTEx: ENSG00000103363 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q15370 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Core-binding factor subunit beta | 170 | Homo sapiens | Mutation(s): 0 Gene Names: CBFB, CBFbeta | 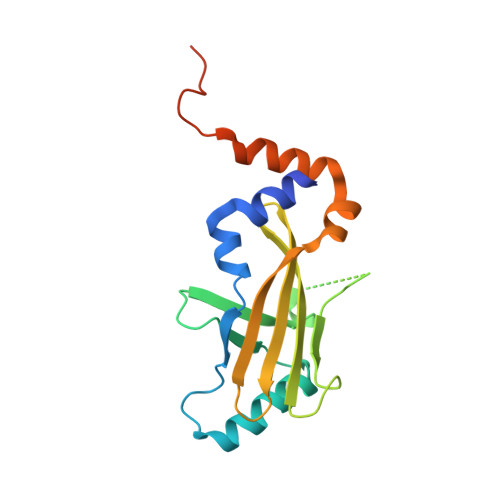 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q13951 (Homo sapiens) Explore Q13951 Go to UniProtKB: Q13951 | |||||
PHAROS: Q13951 GTEx: ENSG00000067955 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q13951 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Virion infectivity factor | 176 | Human immunodeficiency virus 1 | Mutation(s): 0 Gene Names: vif |  | |
UniProt | |||||
Find proteins for P12504 (Human immunodeficiency virus type 1 group M subtype B (isolate NY5)) Explore P12504 Go to UniProtKB: P12504 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P12504 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| ZN Query on ZN | IB [auth b] JB [auth G] KB [auth M] LB [auth S] MB [auth d] | ZINC ION Zn PTFCDOFLOPIGGS-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 103.925 | α = 65.51 |
| b = 204.002 | β = 90.28 |
| c = 247.866 | γ = 90.5 |
| Software Name | Purpose |
|---|---|
| HKL-2000 | data collection |
| PHASES | phasing |
| PHENIX | refinement |
| HKL-2000 | data reduction |
| HKL-2000 | data scaling |