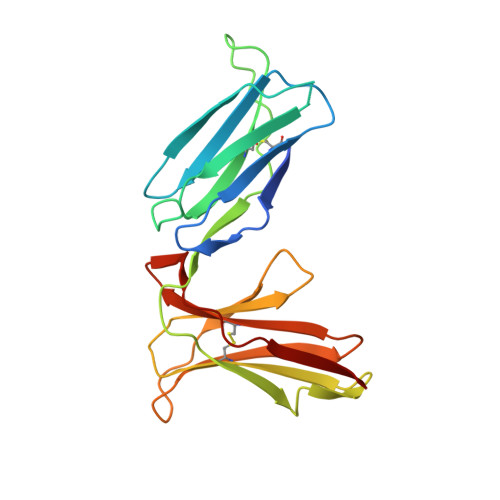
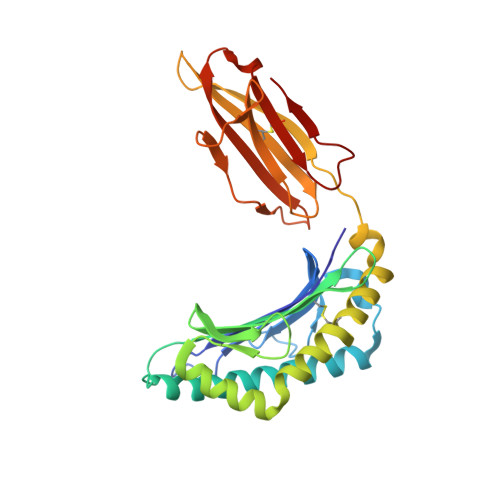
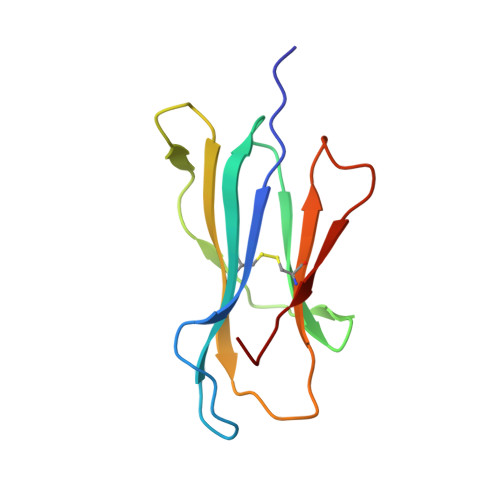
Activating killer cell immunoglobulin-like receptor 2DS2 binds to HLA-A*11
Liu, J.X., Xiao, Z.W., Ko, H.L., Shen, M.X., Ren, E.C.(2014) Proc Natl Acad Sci U S A 111: 2662-2667
- PubMed: 24550293
- DOI: https://doi.org/10.1073/pnas.1322052111
- Primary Citation of Related Structures:
4N8V - PubMed Abstract:
Inhibitory killer cell Ig-like receptors (KIRs) are known to recognize HLA ligands mainly of the HLA-C and Bw4 groups, but the ligands for KIRs are poorly understood. We report here the identification of the cognate ligand for the activating KIR 2DS2 as HLA-A*11:01. The crystal structure of the KIR2DS2-HLA-A*11:01 complex was solved at 2.5-Å resolution and revealed residue-binding characteristics distinct from those of inhibitory KIRs with HLA-C and the critical role of residues Tyr45 and Asp72 in shaping binding specificity to HLA-A*11:01. Using KIR2DS2 tetramers, binding to surface HLA-A*11:01 on live cells was demonstrated and, furthermore, that binding can be altered by residue changes at p8 of the peptide, indicating the influence of peptide sequence on KIR-HLA association. In addition, heteronuclear single quantum coherence NMR was used to map the involvement of critical residues in HLA binding at the interface of KIR and HLA, and validates the data observed in the crystal structure. Our data provide structural evidence of the recognition of A*11:01 by the activating KIR2DS2 and extend our understanding of the KIR-HLA binding spectrum.
- Singapore Immunology Network, Agency for Science, Technology and Research, Singapore 138648.
Organizational Affiliation: