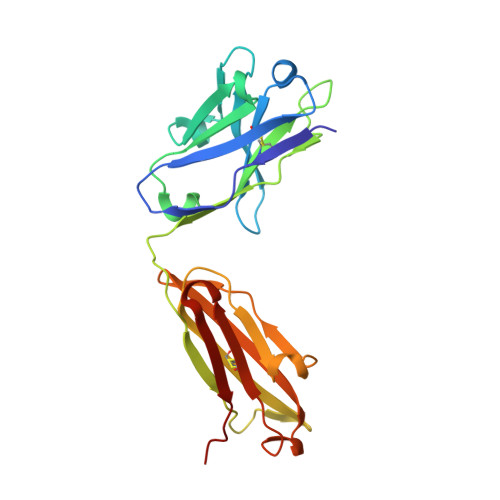
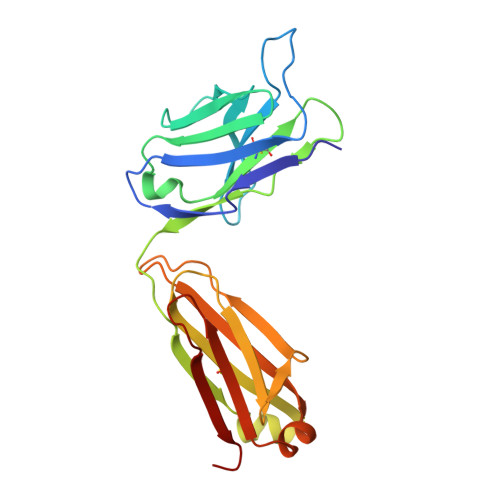
Structure of the extracellular domain of matrix protein 2 of influenza A virus in complex with a protective monoclonal antibody
Cho, K.J., Schepens, B., Seok, J.H., Kim, S., Roose, K., Lee, J.H., Gallardo, R., Hamme, E.V., Schymkowitz, J., Rousseau, F., Fiers, W., Saelens, X., Kim, K.H.(2015) J Virol 89: 3700-3711
- PubMed: 25609808
- DOI: https://doi.org/10.1128/JVI.02576-14
- Primary Citation of Related Structures:
4N8C - PubMed Abstract:
The extracellular domain of influenza A virus matrix protein 2 (M2e) is conserved and is being evaluated as a quasiuniversal influenza A vaccine candidate. We describe the crystal structure at 1.6 Å resolution of M2e in complex with the Fab fragment of an M2e-specific monoclonal antibody that protects against influenza A virus challenge. This antibody binds M2 expressed on the surfaces of cells infected with influenza A virus. Five out of six complementary determining regions interact with M2e, and three highly conserved M2e residues are critical for this interaction. In this complex, M2e adopts a compact U-shaped conformation stabilized in the center by the highly conserved tryptophan residue in M2e. This is the first description of the three-dimensional structure of M2e. M2e of influenza A is under investigation as a universal influenza A vaccine, but its three-dimensional structure is unknown. We describe the structure of M2e stabilized with an M2e-specific monoclonal antibody that recognizes natural M2. We found that the conserved tryptophan is positioned in the center of the U-shaped structure of M2e and stabilizes its conformation. The structure also explains why previously reported in vivo escape viruses, selected with a similar monoclonal antibody, carried proline residue substitutions at position 10 in M2.
- Department of Biotechnology & Bioinformatics, College of Science & Technology, Korea University, Sejong, South Korea Inflammation Research Center, VIB, Ghent, Belgium Department of Biomedical Molecular Biology, Ghent University, Ghent, Belgium.
Organizational Affiliation: