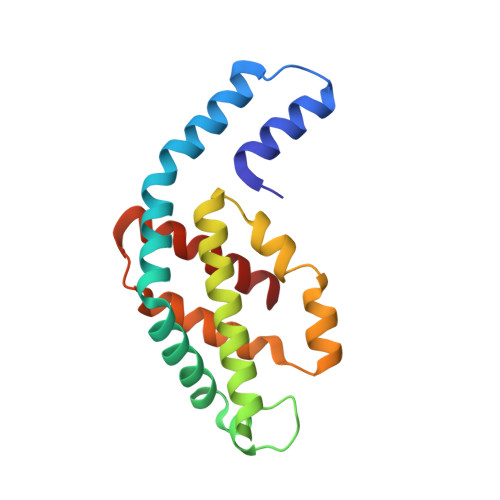
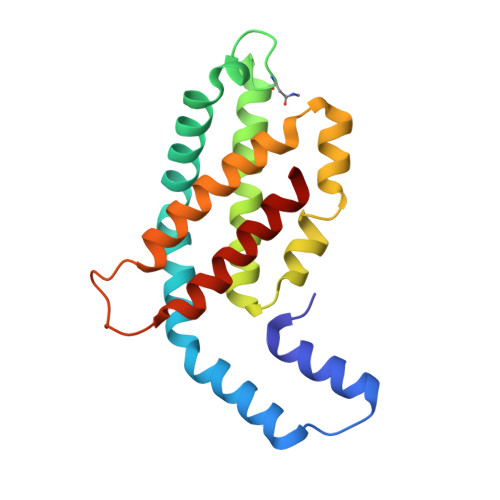
Structural studies show energy transfer within stabilized phycobilisomes independent of the mode of rod-core assembly.
David, L., Prado, M., Arteni, A.A., Elmlund, D.A., Blankenship, R.E., Adir, N.(2014) Biochim Biophys Acta 1837: 385-395
- PubMed: 24407142
- DOI: https://doi.org/10.1016/j.bbabio.2013.12.014
- Primary Citation of Related Structures:
4N6S - PubMed Abstract:
The major light harvesting complex in cyanobacteria and red algae is the phycobilisome (PBS), comprised of hundreds of seemingly similar chromophores, which are protein bound and assembled in a fashion that enables highly efficient uni-directional energy transfer to reaction centers. The PBS is comprised of a core containing 2-5 cylinders surrounded by 6-8 rods, and a number of models have been proposed describing the PBS structure. One of the most critical steps in the functionality of the PBS is energy transfer from the rod substructures to the core substructure. In this study we compare the structural and functional characteristics of high-phosphate stabilized PBS (the standard fashion of stabilization of isolated complexes) with cross-linked PBS in low ionic strength buffer from two cyanobacterial species, Thermosynechococcus vulcanus and Acaryochloris marina. We show that chemical cross-linking preserves efficient energy transfer from the phycocyanin containing rods to the allophycocyanin containing cores with fluorescent emission from the terminal emitters. However, this energy transfer is shown to exist in PBS complexes of different structures as characterized by determination of a 2.4Å structure by X-ray crystallography, single crystal confocal microscopy, mass spectrometry and transmission electron microscopy of negatively stained and cryogenically preserved complexes. We conclude that the PBS has intrinsic structural properties that enable efficient energy transfer from rod substructures to the core substructures without requiring a single unique structure. We discuss the significance of our observations on the functionality of the PBS in vivo.
- Schulich Faculty of Chemistry, Technion-Israel Institute of Technology, Haifa 32000, Israel.
Organizational Affiliation: