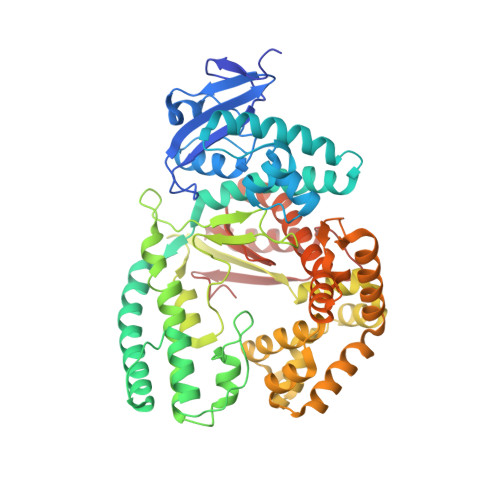
A Conservative Isoleucine to Leucine Mutation Causes Major Rearrangements and Cold Sensitivity in KlenTaq1 DNA Polymerase.
Wu, E.Y., Walsh, A.R., Materne, E.C., Hiltner, E.P., Zielinski, B., Miller, B.R., Mawby, L., Modeste, E., Parish, C.A., Barnes, W.M., Kermekchiev, M.B.(2015) Biochemistry 54: 881-889
- PubMed: 25537790
- DOI: https://doi.org/10.1021/bi501198f
- Primary Citation of Related Structures:
4N56, 4N5S, 4XIU - PubMed Abstract:
Assembly of polymerase chain reactions at room temperature can sometimes lead to low yields or unintentional products due to mispriming. Mutation of isoleucine 707 to leucine in DNA polymerase I from Thermus aquaticus substantially decreases its activity at room temperature without compromising its ability to amplify DNA. To understand why a conservative change to the enzyme over 20 Å from the active site can have a large impact on its activity at low temperature, we solved the X-ray crystal structure of the large (5'-to-3' exonuclease-deleted) fragment of Taq DNA polymerase containing the cold-sensitive mutation in the ternary (E-DNA-ddNTP) and binary (E-DNA) complexes. The I707L KlenTaq1 ternary complex was identical to the wild-type in the closed conformation except for the mutation and a rotamer change in nearby phenylalanine 749, suggesting that the enzyme should remain active. However, soaking out of the nucleotide substrate at low temperature results in an altered binary complex made possible by the rotamer change at F749 near the tip of the polymerase O-helix. Surprisingly, two adenosines in the 5'-template overhang fill the vacated active site by stacking with the primer strand, thereby blocking the active site at low temperature. Replacement of the two overhanging adenosines with pyrimidines substantially increased activity at room temperature by keeping the template overhang out of the active site, confirming the importance of base stacking. These results explain the cold-sensitive phenotype of the I707L mutation in KlenTaq1 and serve as an example of a large conformational change affected by a conservative mutation.
- Department of Biology and ‡Department of Chemistry, University of Richmond , Richmond, Virginia 23173, United States.
Organizational Affiliation: