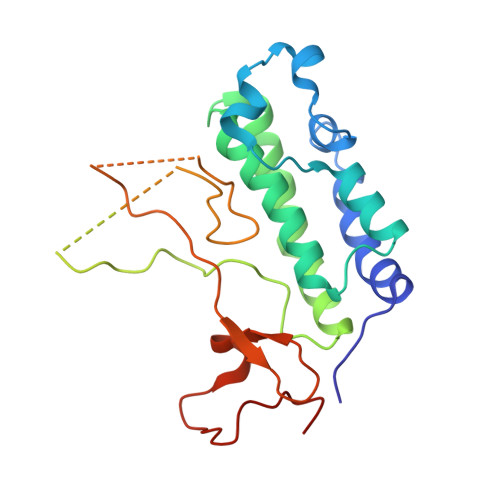
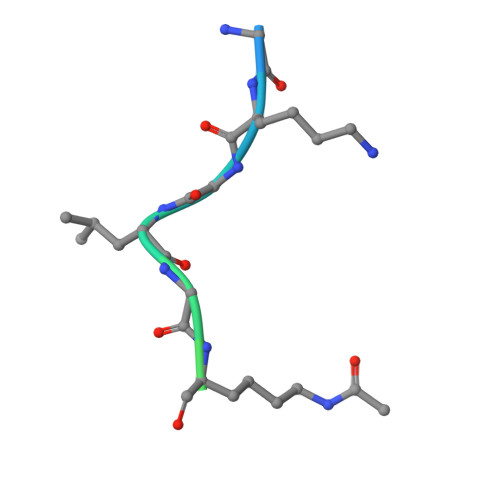
Structural Insights into Acetylated-Histone H4 Recognition by the Bromodomain-PHD Finger Module of Human Transcriptional Coactivator CBP.
Plotnikov, A.N., Yang, S., Zhou, T.J., Rusinova, E., Frasca, A., Zhou, M.M.(2014) Structure 22: 353-360
- PubMed: 24361270
- DOI: https://doi.org/10.1016/j.str.2013.10.021
- Primary Citation of Related Structures:
4N3W, 4N4F - PubMed Abstract:
Bromodomain functions as the acetyl-lysine binding domains to regulate gene transcription in chromatin. Bromodomains are rapidly emerging as new epigenetic drug targets for human diseases. However, owing to their transient nature and modest affinity, histone-binding selectivity of bromodomains has remained mostly elusive. Here, we report high-resolution crystal structures of the bromodomain-PHD tandem module of human transcriptional coactivator CBP bound to lysine-acetylated histone H4 peptides. The structures reveal that the PHD finger serves a structural role in the tandem module and that the bromodomain prefers lysine-acetylated motifs comprising a hydrophobic or aromatic residue at -2 and a lysine or arginine at -3 or -4 position from the acetylated lysine. Our study further provides structural insights into distinct modes of singly and diacetylated histone H4 recognition by the bromodomains of CBP and BRD4 that function differently as a transcriptional coactivator and chromatin organizer, respectively, explaining their distinct roles in control of gene expression in chromatin.
- Department of Structural and Chemical Biology, Icahn School of Medicine at Mount Sinai, 1425 Madison Avenue, New York, NY 10029, USA.
Organizational Affiliation: