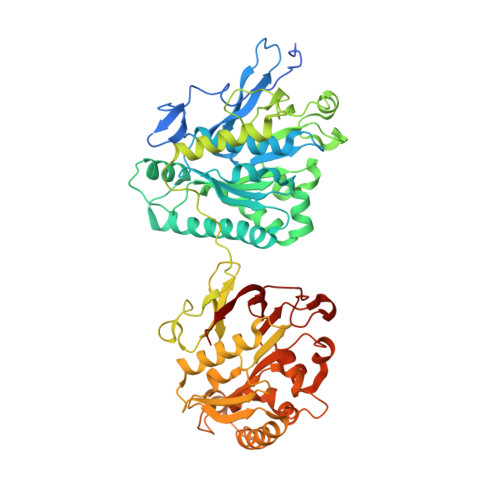
Substrates Control Multimerization and Activation of the Multi-Domain ATPase Motor of Type VII Secretion.
Rosenberg, O.S., Dovala, D., Li, X., Connolly, L., Bendebury, A., Finer-Moore, J., Holton, J., Cheng, Y., Stroud, R.M., Cox, J.S.(2015) Cell 161: 501-512
- PubMed: 25865481
- DOI: https://doi.org/10.1016/j.cell.2015.03.040
- Primary Citation of Related Structures:
4LWS, 4LYA, 4N1A, 4NH0 - PubMed Abstract:
Mycobacterium tuberculosis and Staphylococcus aureus secrete virulence factors via type VII protein secretion (T7S), a system that intriguingly requires all of its secretion substrates for activity. To gain insights into T7S function, we used structural approaches to guide studies of the putative translocase EccC, a unique enzyme with three ATPase domains, and its secretion substrate EsxB. The crystal structure of EccC revealed that the ATPase domains are joined by linker/pocket interactions that modulate its enzymatic activity. EsxB binds via its signal sequence to an empty pocket on the C-terminal ATPase domain, which is accompanied by an increase in ATPase activity. Surprisingly, substrate binding does not activate EccC allosterically but, rather, by stimulating its multimerization. Thus, the EsxB substrate is also an integral T7S component, illuminating a mechanism that helps to explain interdependence of substrates, and suggests a model in which binding of substrates modulates their coordinate release from the bacterium.
- Division of Infectious Diseases, Department of Medicine, UCSF Medical Center, University of California, San Francisco, San Francisco, CA 94143-0654, USA.
Organizational Affiliation: