TBN
Wang, J., Zajonc, D.M.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Antigen-presenting glycoprotein CD1d1 | 285 | Mus musculus | Mutation(s): 0 Gene Names: Cd1.1, Cd1d1, mcd1d | 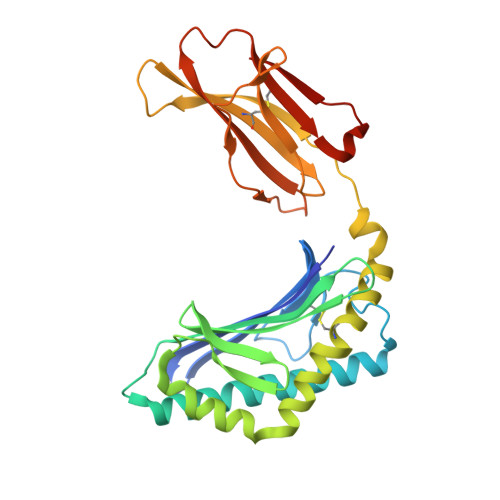 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P11609 (Mus musculus) Explore P11609 Go to UniProtKB: P11609 | |||||
IMPC: MGI:107674 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P11609 | ||||
Glycosylation | |||||
| Glycosylation Sites: 3 | Go to GlyGen: P11609-1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Beta-2-microglobulin | 99 | Mus musculus | Mutation(s): 0 Gene Names: B2m | 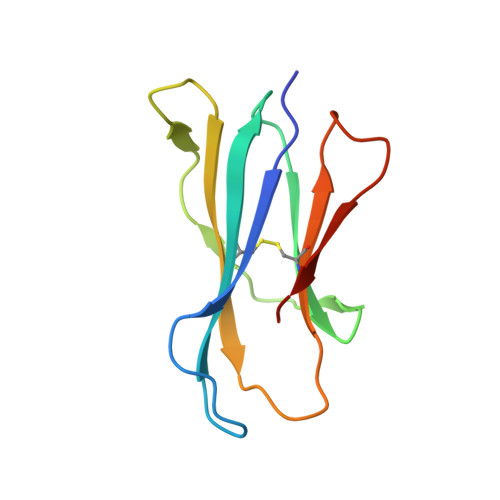 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P01887 (Mus musculus) Explore P01887 Go to UniProtKB: P01887 | |||||
IMPC: MGI:88127 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P01887 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| MX7 Query on MX7 | F [auth A] | (2R)-3-(phosphonooxy)propane-1,2-diyl (9Z,9'Z)bis-octadec-9-enoate C39 H73 O8 P MHUWZNTUIIFHAS-DSSVUWSHSA-N |  | ||
| NAG Query on NAG | D [auth A], E [auth A] | 2-acetamido-2-deoxy-beta-D-glucopyranose C8 H15 N O6 OVRNDRQMDRJTHS-FMDGEEDCSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 42.2 | α = 90 |
| b = 106.3 | β = 90 |
| c = 107.48 | γ = 90 |
| Software Name | Purpose |
|---|---|
| Blu-Ice | data collection |
| MOLREP | phasing |
| REFMAC | refinement |
| MOSFLM | data reduction |
| SCALA | data scaling |