Development and evaluation of a sublingual tablet based on recombinant Bet v 1 in birch pollen-allergic patients.
Nony, E., Bouley, J., Le Mignon, M., Lemoine, P., Jain, K., Horiot, S., Mascarell, L., Pallardy, M., Vincentelli, R., Leone, P., Roussel, A., Batard, T., Abiteboul, K., Robin, B., de Beaumont, O., Arvidsson, M., Rak, S., Moingeon, P.(2015) Allergy 70: 795-804
- PubMed: 25846209
- DOI: https://doi.org/10.1111/all.12622
- Primary Citation of Related Structures:
4MNS - PubMed Abstract:
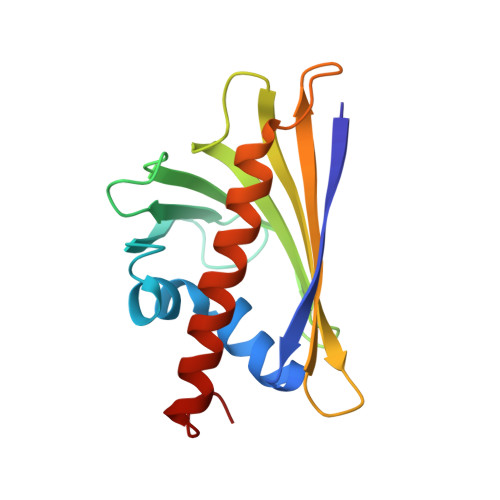
Sublingual immunotherapy (SLIT) applied to type I respiratory allergies is commonly performed with natural allergen extracts. Herein, we developed a sublingual tablet made of pharmaceutical-grade recombinant Bet v 1.0101 (rBet v 1) and investigated its clinical safety and efficacy in birch pollen (BP)-allergic patients. Following expression in Escherichia coli and purification, rBet v 1 was characterized using chromatography, capillary electrophoresis, circular dichroism, mass spectrometry and crystallography. Safety and efficacy of rBet v 1 formulated as a sublingual tablet were assessed in a multicentre, double-blind, placebo-controlled study conducted in 483 patients with BP-induced rhinoconjunctivitis. In-depth characterization confirmed the intact product structure and high purity of GMP-grade rBet v 1. The crystal structure resolved at 1.2 Å documented the natural conformation of the molecule. Native or oxidized forms of rBet v 1 did not induce the production of any proinflammatory cytokine by blood dendritic cells or mononuclear cells. Bet v 1 tablets were well tolerated by patients, consistent with the known safety profile of SLIT. The average adjusted symptom scores were significantly decreased relative to placebo in patients receiving once daily for 5 months rBet v 1 tablets, with a mean difference of 17.0-17.7% relative to the group treated with placebo (P < 0.025), without any influence of the dose in the range (12.5-50 μg) tested. Recombinant Bet v 1 has been produced as a well-characterized pharmaceutical-grade biological drug. Sublingual administration of rBet v 1 tablets is safe and efficacious in patients with BP allergic rhinoconjunctivitis.
- Stallergenes, Antony, France.
Organizational Affiliation: