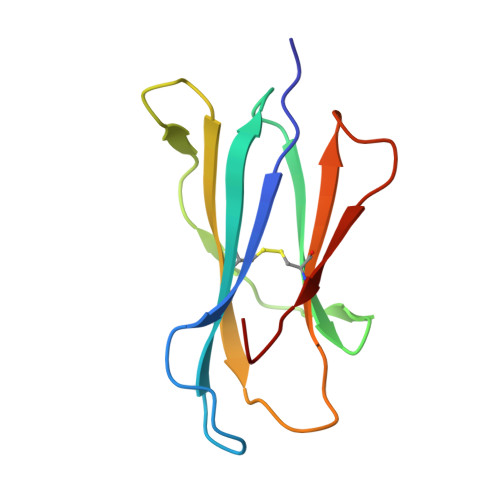
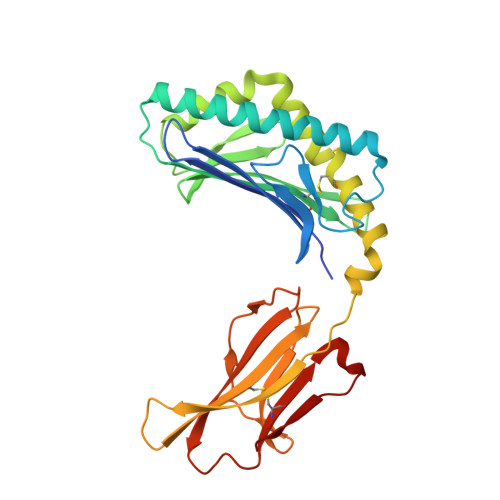
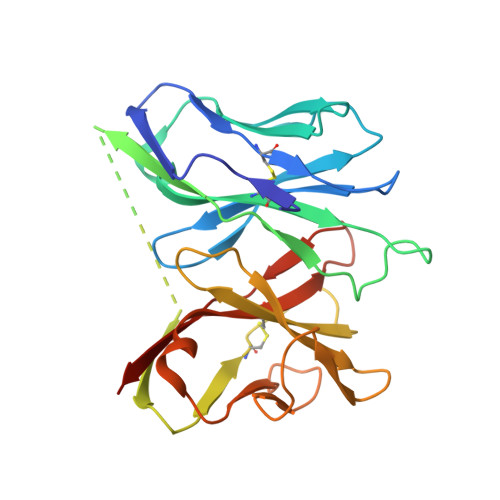
Crystal Structure of V delta 1 T Cell Receptor in Complex with CD1d-Sulfatide Shows MHC-like Recognition of a Self-Lipid by Human gamma delta T Cells.
Luoma, A.M., Castro, C.D., Mayassi, T., Bembinster, L.A., Bai, L., Picard, D., Anderson, B., Scharf, L., Kung, J.E., Sibener, L.V., Savage, P.B., Jabri, B., Bendelac, A., Adams, E.J.(2013) Immunity 39: 1032-1042
- PubMed: 24239091
- DOI: https://doi.org/10.1016/j.immuni.2013.11.001
- Primary Citation of Related Structures:
4MNG, 4MNH, 4MQ7, 4NDM - PubMed Abstract:
The nature of the antigens recognized by γδ T cells and their potential recognition of major histocompatibility complex (MHC)-like molecules has remained unclear. Members of the CD1 family of lipid-presenting molecules are suggested ligands for Vδ1 TCR-expressing γδ T cells, the major γδ lymphocyte population in epithelial tissues. We crystallized a Vδ1 TCR in complex with CD1d and the self-lipid sulfatide, revealing the unusual recognition of CD1d by germline Vδ1 residues spanning all complementarity-determining region (CDR) loops, as well as sulfatide recognition separately encoded by nongermline CDR3δ residues. Binding and functional analysis showed that CD1d presenting self-lipids, including sulfatide, was widely recognized by gut Vδ1+ γδ T cells. These findings provide structural demonstration of MHC-like recognition of a self-lipid by γδ T cells and reveal the prevalence of lipid recognition by innate-like T cell populations.
- Committee on Immunology, University of Chicago, Chicago, IL 60637, USA; Department of Biochemistry and Molecular Biology, University of Chicago, Chicago, IL 60637, USA.
Organizational Affiliation: