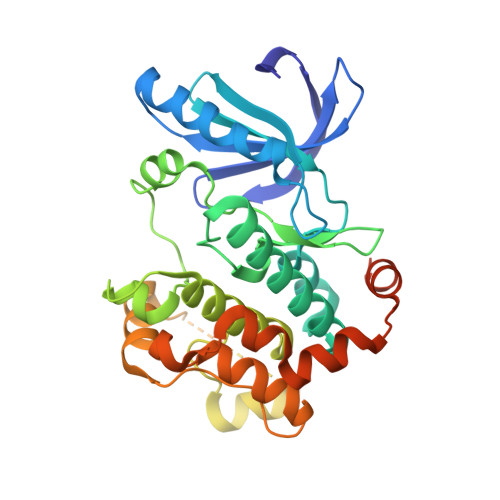
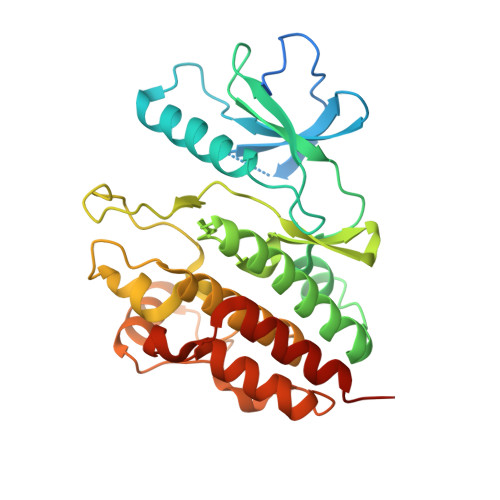
Structure of the BRAF-MEK Complex Reveals a Kinase Activity Independent Role for BRAF in MAPK Signaling.
Haling, J.R., Sudhamsu, J., Yen, I., Sideris, S., Sandoval, W., Phung, W., Bravo, B.J., Giannetti, A.M., Peck, A., Masselot, A., Morales, T., Smith, D., Brandhuber, B.J., Hymowitz, S.G., Malek, S.(2014) Cancer Cell 26: 402-413
- PubMed: 25155755
- DOI: https://doi.org/10.1016/j.ccr.2014.07.007
- Primary Citation of Related Structures:
4MNE, 4MNF - PubMed Abstract:
Numerous oncogenic mutations occur within the BRAF kinase domain (BRAF(KD)). Here we show that stable BRAF-MEK1 complexes are enriched in BRAF(WT) and KRAS mutant (MT) cells but not in BRAF(MT) cells. The crystal structure of the BRAF(KD) in a complex with MEK1 reveals a face-to-face dimer sensitive to MEK1 phosphorylation but insensitive to BRAF dimerization. Structure-guided studies reveal that oncogenic BRAF mutations function by bypassing the requirement for BRAF dimerization for activity or weakening the interaction with MEK1. Finally, we show that conformation-specific BRAF inhibitors can sequester a dormant BRAF-MEK1 complex resulting in pathway inhibition. Taken together, these findings reveal a regulatory role for BRAF in the MAPK pathway independent of its kinase activity but dependent on interaction with MEK.
- Department of Biochemical and Cellular Pharmacology, Genentech Inc., 1 DNA Way, South San Francisco, CA 94080, USA.
Organizational Affiliation: