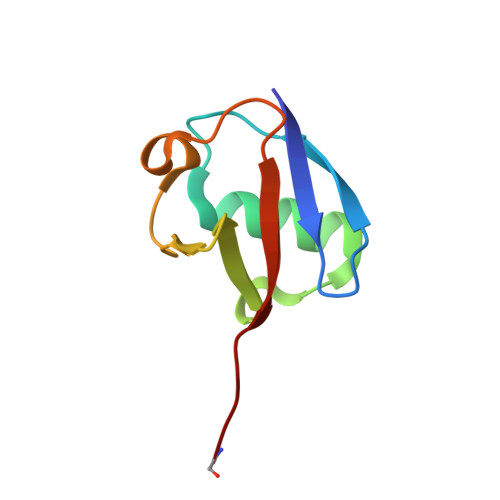
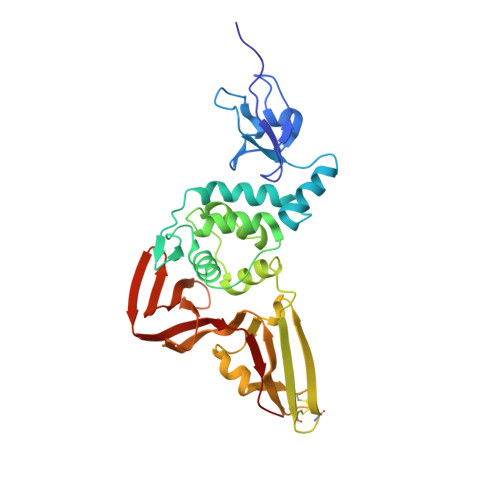
Structural Basis for the Ubiquitin-Linkage Specificity and deISGylating activity of SARS-CoV papain-like protease.
Ratia, K., Kilianski, A., Baez-Santos, Y.M., Baker, S.C., Mesecar, A.(2014) PLoS Pathog 10: e1004113-e1004113
- PubMed: 24854014
- DOI: https://doi.org/10.1371/journal.ppat.1004113
- Primary Citation of Related Structures:
4MM3 - PubMed Abstract:
Severe acute respiratory syndrome coronavirus (SARS-CoV) encodes a papain-like protease (PLpro) with both deubiquitinating (DUB) and deISGylating activities that are proposed to counteract the post-translational modification of signaling molecules that activate the innate immune response. Here we examine the structural basis for PLpro's ubiquitin chain and interferon stimulated gene 15 (ISG15) specificity. We present the X-ray crystal structure of PLpro in complex with ubiquitin-aldehyde and model the interaction of PLpro with other ubiquitin-chain and ISG15 substrates. We show that PLpro greatly prefers K48- to K63-linked ubiquitin chains, and ISG15-based substrates to those that are mono-ubiquitinated. We propose that PLpro's higher affinity for K48-linked ubiquitin chains and ISG15 stems from a bivalent mechanism of binding, where two ubiquitin-like domains prefer to bind in the palm domain of PLpro with the most distal ubiquitin domain interacting with a "ridge" region of the thumb domain. Mutagenesis of residues within this ridge region revealed that these mutants retain viral protease activity and the ability to catalyze hydrolysis of mono-ubiquitin. However, a select number of these mutants have a significantly reduced ability to hydrolyze the substrate ISG15-AMC, or be inhibited by K48-linked diubuiquitin. For these latter residues, we found that PLpro antagonism of the nuclear factor kappa-light-chain-enhancer of activated B-cells (NFκB) signaling pathway is abrogated. This identification of key and unique sites in PLpro required for recognition and processing of diubiquitin and ISG15 versus mono-ubiquitin and protease activity provides new insight into ubiquitin-chain and ISG15 recognition and highlights a role for PLpro DUB and deISGylase activity in antagonism of the innate immune response.
- Department of Medicinal Chemistry and Pharmacognosy, University of Illinois, Chicago, Illinois, United States of America.
Organizational Affiliation: