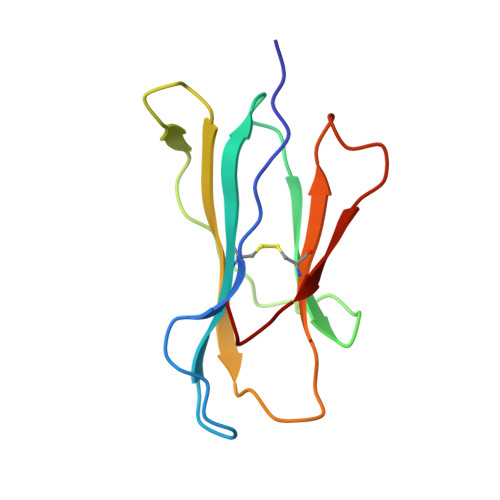
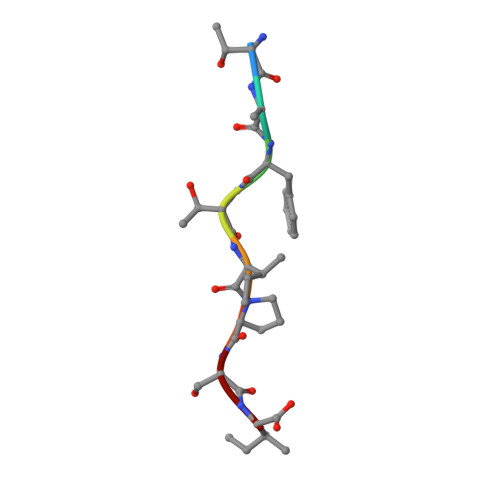
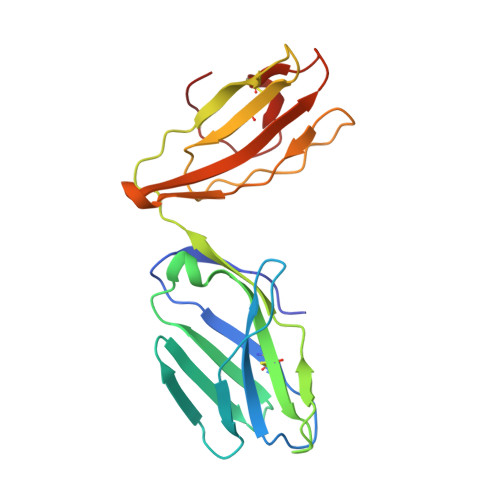
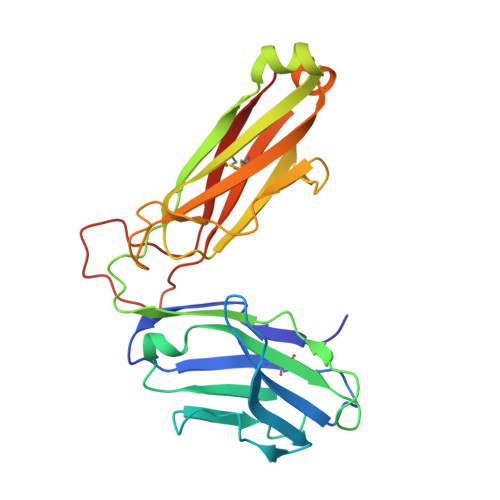
Molecular basis of a dominant T cell response to an HIV reverse transcriptase 8-mer epitope presented by the protective allele HLA-B*51:01
Motozono, C., Kuse, N., Sun, X., Rizkallah, P.J., Fuller, A., Oka, S., Cole, D.K., Sewell, A.K., Takiguchi, M.(2014) J Immunol 192: 3428-3434
- PubMed: 24600035
- DOI: https://doi.org/10.4049/jimmunol.1302667
- Primary Citation of Related Structures:
4MJI - PubMed Abstract:
CD8(+) CTL responses directed toward the HLA-B*51:01-restricted HIV-RT128-135 epitope TAFTIPSI (TI8) are associated with long-term nonprogression to AIDS. Clonotypic analysis of responses to B51-TI8 revealed a public clonotype using TRAV17/TRBV7-3 TCR genes in six out of seven HLA-B*51:01(+) patients. Structural analysis of a TRAV17/TRBV7-3 TCR in complex with HLA-B51-TI8, to our knowledge the first human TCR complexed with an 8-mer peptide, explained this bias, as the unique combination of residues encoded by these genes was central to the interaction. The relatively featureless peptide-MHC (pMHC) was mainly recognized by the TCR CDR1 and CDR2 loops in an MHC-centric manner. A highly conserved residue Arg(97) in the CDR3α loop played a major role in recognition of peptide and MHC to form a stabilizing ball-and-socket interaction with the MHC and peptide, contributing to the selection of the public TCR clonotype. Surface plasmon resonance equilibrium binding analysis showed the low affinity of this public TCR is in accordance with the only other 8-mer interaction studied to date (murine 2C TCR-H-2K(b)-dEV8). Like pMHC class II complexes, 8-mer peptides do not protrude out the MHC class I binding groove like those of longer peptides. The accumulated evidence suggests that weak affinity might be a common characteristic of TCR binding to featureless pMHC landscapes.
- Cardiff University School of Medicine, Heath Park CF14 4XN, United Kingdom;
Organizational Affiliation: