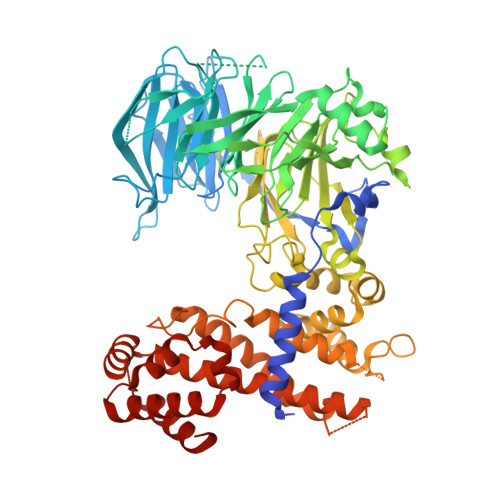
Structure and nucleic acid binding activity of the nucleoporin Nup157.
Seo, H.S., Blus, B.J., Jankovic, N.Z., Blobel, G.(2013) Proc Natl Acad Sci U S A 110: 16450-16455
- PubMed: 24062435
- DOI: https://doi.org/10.1073/pnas.1316607110
- Primary Citation of Related Structures:
4MHC - PubMed Abstract:
At the center of the nuclear pore complex (NPC) is a uniquely versatile central transport channel. Structural analyses of distinct segments ("protomers") of the three "channel" nucleoporins yielded a model for how this channel is constructed. Its principal feature is a midplane ring that can undergo regulated diameter changes of as much as an estimated 30 nm. To better understand how a family of "adaptor" nucleoporins--concentrically surrounding this channel--might cushion these huge structural changes, we determined the crystal structure of one adaptor nucleoporin, Nup157. Here, we show that a recombinant Saccharomyces cerevisiae Nup157 protomer, representing two-thirds of Nup157 (residues 70-893), folds into a seven-bladed β-propeller followed by an α-helical domain, which together form a C-shaped architecture. Notably, the structure contains a large patch of positively charged residues, most of which are evolutionarily conserved. Consistent with this surface feature, we found that Nup157(70-893) binds to nucleic acids, although in a sequence-independent manner. Nevertheless, this interaction supports a previously reported role of Nup157, and its paralogue Nup170, in chromatin organization. Based on its nucleic acid binding capacity, we propose a dual location and function of Nup157. Finally, modeling the remaining C-terminal portion of Nup157 shows that it projects as a superhelical stack from the compact C-shaped portion of the molecule. The predicted four hinge regions indicate an intrinsic flexibility of Nup157, which could contribute to structural plasticity within the NPC.
- Laboratory of Cell Biology and Howard Hughes Medical Institute, The Rockefeller University, New York, NY 10065.
Organizational Affiliation: