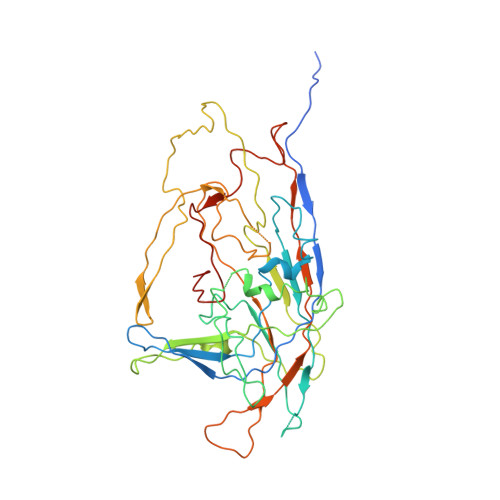
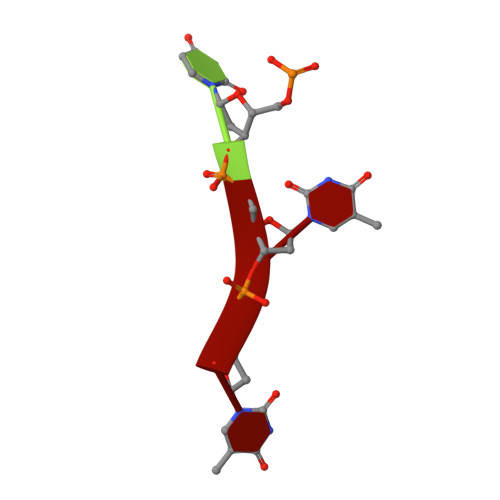
The structure and host entry of an invertebrate parvovirus.
Meng, G., Zhang, X., Plevka, P., Yu, Q., Tijssen, P., Rossmann, M.G.(2013) J Virol 87: 12523-12530
- PubMed: 24027306
- DOI: https://doi.org/10.1128/JVI.01822-13
- Primary Citation of Related Structures:
4MGU - PubMed Abstract:
The 3.5-Å resolution X-ray crystal structure of mature cricket parvovirus (Acheta domesticus densovirus [AdDNV]) has been determined. Structural comparisons show that vertebrate and invertebrate parvoviruses have evolved independently, although there are common structural features among all parvovirus capsid proteins. It was shown that raising the temperature of the AdDNV particles caused a loss of their genomes. The structure of these emptied particles was determined by cryo-electron microscopy to 5.5-Å resolution, and the capsid structure was found to be the same as that for the full, mature virus except for the absence of the three ordered nucleotides observed in the crystal structure. The viral protein 1 (VP1) amino termini could be externalized without significant damage to the capsid. In vitro, this externalization of the VP1 amino termini is accompanied by the release of the viral genome.
- Department of Biological Sciences, Purdue University, West Lafayette, Indiana, USA.
Organizational Affiliation: