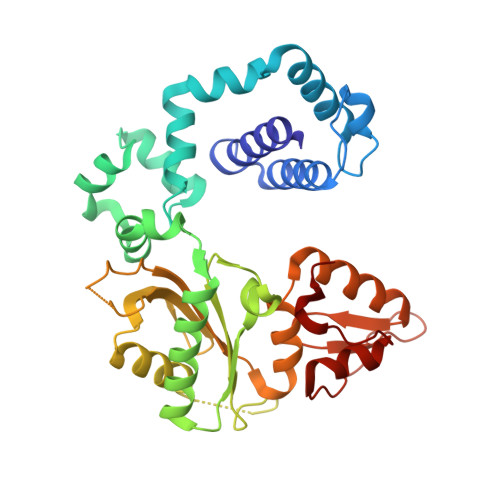
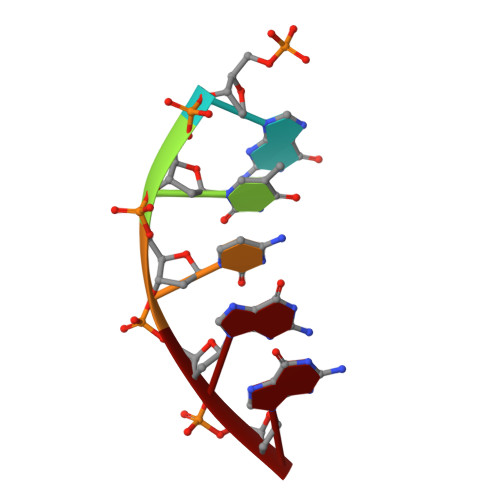
Metal-dependent conformational activation explains highly promutagenic replication across O6-methylguanine by human DNA polymerase beta.
Koag, M.C., Lee, S.(2014) J Am Chem Soc 136: 5709-5721
- PubMed: 24694247
- DOI: https://doi.org/10.1021/ja500172d
- Primary Citation of Related Structures:
4MF2, 4MFC, 4MFF, 4NXZ, 4NY8 - PubMed Abstract:
Human DNA polymerase β (polβ) inserts, albeit slowly, T opposite the carcinogenic lesion O6-methylguanine (O6MeG) ∼30-fold more frequently than C. To gain insight into this promutagenic process, we solved four ternary structures of polβ with an incoming dCTP or dTTP analogue base-paired with O6MeG in the presence of active-site Mg(2+) or Mn(2+). The Mg(2+)-bound structures show that both the O6MeG·dCTP/dTTP-Mg(2+) complexes adopt an open protein conformation, staggered base pair, and one active-site metal ion. The Mn(2+)-bound structures reveal that, whereas the O6Me·dCTP-Mn(2+) complex assumes the similar altered conformation, the O6MeG·dTTP-Mn(2+) complex adopts a catalytically competent state with a closed protein conformation and pseudo-Watson-Crick base pair. On the basis of these observations, we conclude that polβ slows nucleotide incorporation opposite O6MeG by inducing an altered conformation suboptimal for catalysis and promotes mutagenic replication by allowing Watson-Crick-mode for O6MeG·T but not for O6MeG·C in the enzyme active site. The O6MeG·dTTP-Mn(2+) ternary structure, which represents the first structure of mismatched polβ ternary complex with a closed protein conformation and coplanar base pair, the first structure of pseudo-Watson-Crick O6MeG·T formed in the active site of a DNA polymerase, and a rare, if not the first, example of metal-dependent conformational activation of a DNA polymerase, indicate that catalytic metal-ion coordination is utilized as a kinetic checkpoint by polβ and is crucial for the conformational activation of polβ. Overall, our structural studies not only explain the promutagenic polβ catalysis across O6MeG but also provide new insights into the replication fidelity of polβ.
- Division of Medicinal Chemistry, College of Pharmacy, The University of Texas at Austin , Austin, Texas 78712, United States.
Organizational Affiliation: