Crystal structure of a putative ssRNA endonuclease Cas2, CRISPR adaptation protein from E.coli
Nocek, B., Skarina, T., Brown, G., Yakunin, A., Joachimiak, A.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| CRISPR-associated endoribonuclease Cas2 | 94 | Escherichia coli str. K-12 substr. MG1655 | Mutation(s): 0 Gene Names: ygbF, cas2, b2754, JW5438 EC: 3.1 | 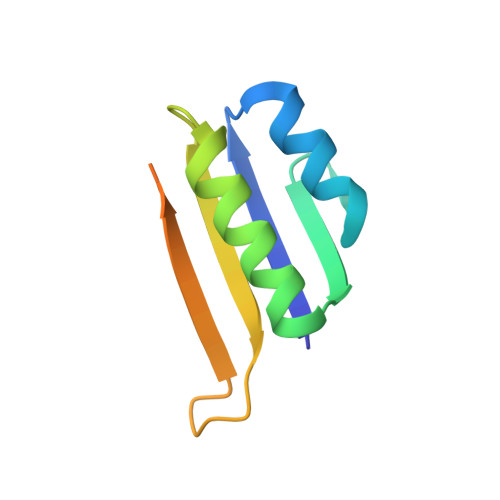 | |
UniProt | |||||
Find proteins for P45956 (Escherichia coli (strain K12)) Explore P45956 Go to UniProtKB: P45956 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P45956 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| PEG Query on PEG | C [auth B] | DI(HYDROXYETHYL)ETHER C4 H10 O3 MTHSVFCYNBDYFN-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 30.048 | α = 90 |
| b = 66.009 | β = 90 |
| c = 69.972 | γ = 90 |
| Software Name | Purpose |
|---|---|
| SBC-Collect | data collection |
| CCP4 | model building |
| MOLREP | phasing |
| REFMAC | refinement |
| HKL-3000 | data reduction |
| HKL-3000 | data scaling |
| CCP4 | phasing |