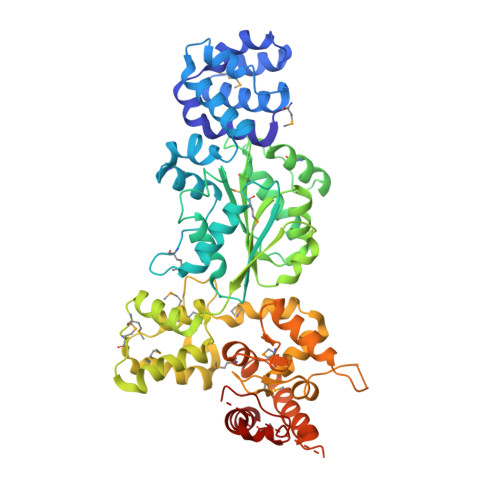
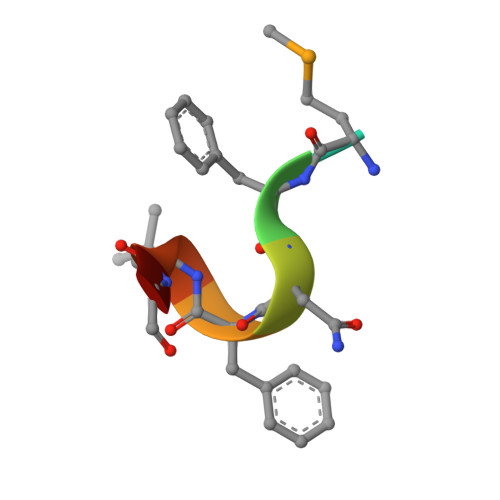
Mechanistic insights into CED-4-mediated activation of CED-3.
Huang, W., Jiang, T., Choi, W., Qi, S., Pang, Y., Hu, Q., Xu, Y., Gong, X., Jeffrey, P.D., Wang, J., Shi, Y.(2013) Genes Dev 27: 2039-2048
- PubMed: 24065769
- DOI: https://doi.org/10.1101/gad.224428.113
- Primary Citation of Related Structures:
4M9R, 4M9S, 4M9X, 4M9Y, 4M9Z - PubMed Abstract:
Programmed cell death in Caenorhabditis elegans requires activation of the caspase CED-3, which strictly depends on CED-4. CED-4 forms an octameric apoptosome, which binds the CED-3 zymogen and facilitates its autocatalytic maturation. Despite recent advances, major questions remain unanswered. Importantly, how CED-4 recognizes CED-3 and how such binding facilitates CED-3 activation remain completely unknown. Here we demonstrate that the L2' loop of CED-3 directly binds CED-4 and plays a major role in the formation of an active CED-4-CED-3 holoenzyme. The crystal structure of the CED-4 apoptosome bound to the L2' loop fragment of CED-3, determined at 3.2 Å resolution, reveals specific interactions between a stretch of five hydrophobic amino acids from CED-3 and a shallow surface pocket within the hutch of the funnel-shaped CED-4 apoptosome. Structure-guided biochemical analysis confirms the functional importance of the observed CED-4-CED-3 interface. Structural analysis together with published evidence strongly suggest a working model in which two molecules of CED-3 zymogen, through specific recognition, are forced into the hutch of the CED-4 apoptosome, consequently undergoing dimerization and autocatalytic maturation. The mechanism of CED-3 activation represents a major revision of the prevailing model for initiator caspase activation.
- Ministry of Education Protein Science Laboratory, Center for Structural Biology, School of Life Sciences.
Organizational Affiliation: