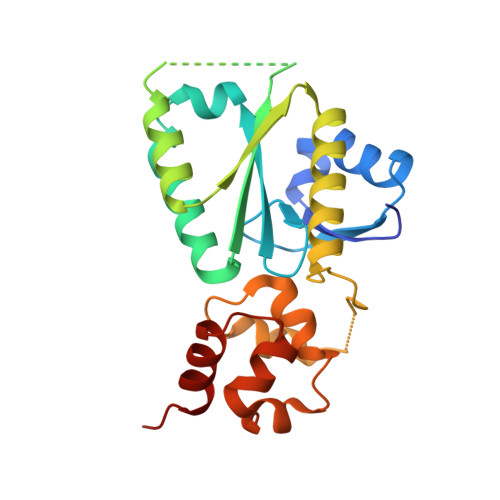
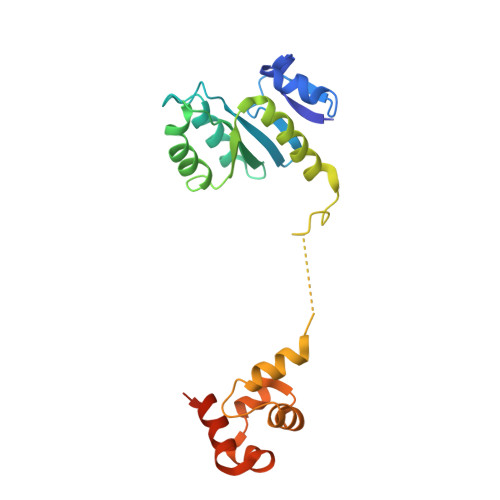Structural insights into the functions of the FANCM-FAAP24 complex in DNA repair.
Yang, H., Zhang, T., Tao, Y., Wang, F., Tong, L., Ding, J.(2013) Nucleic Acids Res 41: 10573-10583
- PubMed: 24003026
- DOI: https://doi.org/10.1093/nar/gkt788
- Primary Citation of Related Structures:
4M6W - PubMed Abstract:
Fanconi anemia (FA) is a genetically heterogeneous disorder associated with deficiencies in the FA complementation group network. FA complementation group M (FANCM) and FA-associated protein 24 kDa (FAAP24) form a stable complex to anchor the FA core complex to chromatin in repairing DNA interstrand crosslinks. Here, we report the first crystal structure of the C-terminal segment of FANCM in complex with FAAP24. The C-terminal segment of FANCM and FAAP24 both consist of a nuclease domain at the N-terminus and a tandem helix-hairpin-helix (HhH)2 domain at the C-terminus. The FANCM-FAAP24 complex exhibits a similar architecture as that of ApXPF. However, the variations of several key residues and the electrostatic property at the active-site region render a catalytically inactive nuclease domain of FANCM, accounting for the lack of nuclease activity. We also show that the first HhH motif of FAAP24 is a potential binding site for DNA, which plays a critical role in targeting FANCM-FAAP24 to chromatin. These results reveal the mechanistic insights into the functions of FANCM-FAAP24 in DNA repair.
- State Key Laboratory of Molecular Biology, Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, 320 Yue-Yang Road, Shanghai 200031, China, Graduate School of Chinese Academy of Sciences, 320 Yue-Yang Road, Shanghai 200031, China and Department of Biological Sciences, Columbia University, New York, NY 10027, USA.
Organizational Affiliation: