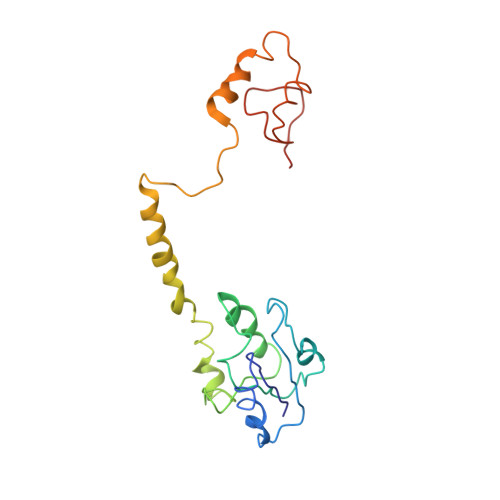
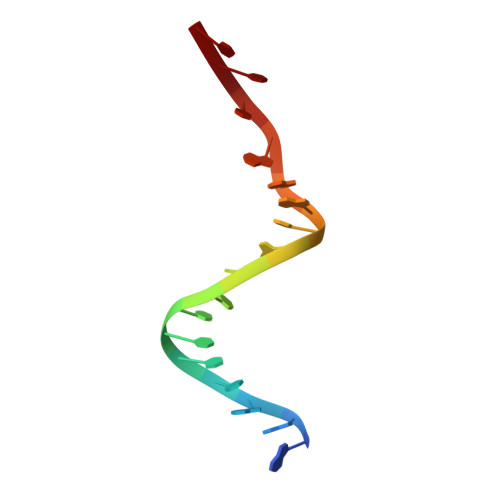
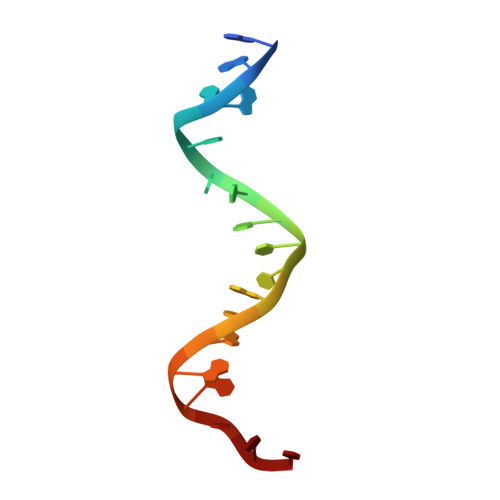
Exploiting large non-isomorphous differences for phase determination of a G-segment invertase-DNA complex.
Ritacco, C.J., Steitz, T.A., Wang, J.(2014) Acta Crystallogr D Biol Crystallogr 70: 685-693
- PubMed: 24598738
- DOI: https://doi.org/10.1107/S1399004713032392
- Primary Citation of Related Structures:
4M6F - PubMed Abstract:
Crystals of the G-segment invertase in complex with a 37-base-pair asymmetric DNA duplex substrate had an unusually high solvent content of 88% and diffracted to a maximal resolution of about 5.0 Å. These crystals exhibited a high degree of non-isomorphism and anisotropy, which presented a serious challenge for structure determination by isomorphous replacement. Here, a procedure of cross-crystal averaging is described that uses large non-isomorphous crystallographic data with a priori information of an approximate molecular boundary as determined from a minimal amount of experimental phase information. Using this procedure, high-quality experimental phases were obtained that have enabled it to be shown that the conformation of the bound substrate DNA duplex significantly differs from those of substrates bound in other serine recombinase-DNA complexes.
- Department of Molecular Biophysics and Biochemistry, Yale University, New Haven, CT 06520, USA.
Organizational Affiliation: