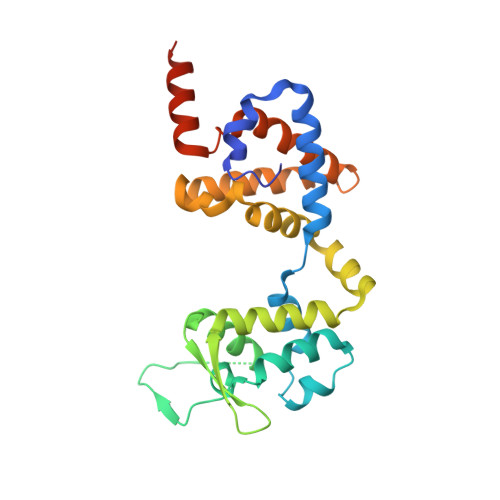
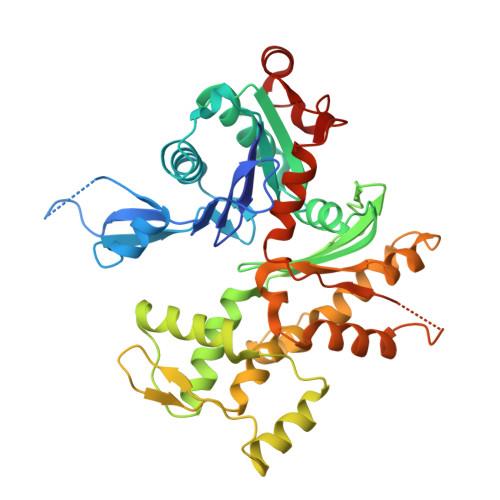
The Bacterial Effector VopL Organizes Actin into Filament-like Structures.
Zahm, J.A., Padrick, S.B., Chen, Z., Pak, C.W., Yunus, A.A., Henry, L., Tomchick, D.R., Chen, Z., Rosen, M.K.(2013) Cell 155: 423-434
- PubMed: 24120140
- DOI: https://doi.org/10.1016/j.cell.2013.09.019
- Primary Citation of Related Structures:
4M63 - PubMed Abstract:
VopL is an effector protein from Vibrio parahaemolyticus that nucleates actin filaments. VopL consists of a VopL C-terminal domain (VCD) and an array of three WASP homology 2 (WH2) motifs. Here, we report the crystal structure of the VCD dimer bound to actin. The VCD organizes three actin monomers in a spatial arrangement close to that found in the canonical actin filament. In this arrangement, WH2 motifs can be modeled into the binding site of each actin without steric clashes. The data suggest a mechanism of nucleation wherein VopL creates filament-like structures, organized by the VCD with monomers delivered by the WH2 array, that can template addition of new subunits. Similarities with Arp2/3 complex and formin proteins suggest that organization of monomers into filament-like structures is a general and central feature of actin nucleation.
- Department of Biophysics, University of Texas Southwestern Medical Center, Dallas, TX 75390, USA; Howard Hughes Medical Institute, University of Texas Southwestern Medical Center, Dallas, TX 75390, USA.
Organizational Affiliation: