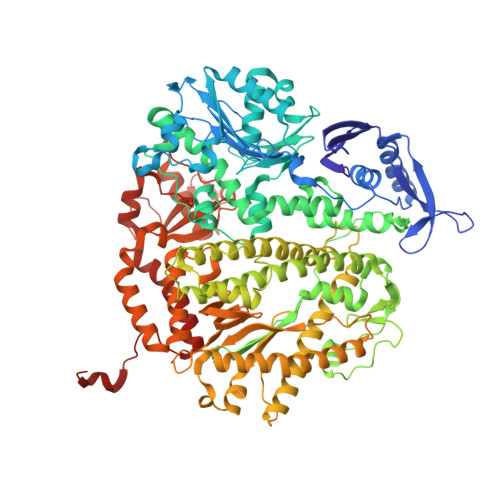
Mispairs with Watson-Crick base-pair geometry observed in ternary complexes of an RB69 DNA polymerase variant.
Xia, S., Konigsberg, W.H.(2014) Protein Sci 23: 508-513
- PubMed: 24458997
- DOI: https://doi.org/10.1002/pro.2434
- Primary Citation of Related Structures:
4M3R, 4M3T, 4M3U, 4M3W, 4M3X, 4M3Y, 4M3Z, 4M41, 4M42, 4M45 - PubMed Abstract:
Recent structures of DNA polymerase complexes with dGMPCPP/dT and dCTP/dA mispairs at the insertion site have shown that they adopt Watson-Crick geometry in the presence of Mn(2+) indicating that the tautomeric or ionization state of the base has changed. To see whether the tautomeric or ionization state of base-pair could be affected by its microenvironment, we determined 10 structures of an RB69 DNA polymerase quadruple mutant with dG/dT or dT/dG mispairs at position n-1 to n-5 of the Primer/Template duplex. Different shapes of the mispairs, including Watson-Crick geometry, have been observed, strongly suggesting that the local environment of base-pairs plays an important role in their tautomeric or ionization states.
- Department of Molecular Biophysics and Biochemistry, Yale University, New Haven, Connecticut, 06520-8114.
Organizational Affiliation: