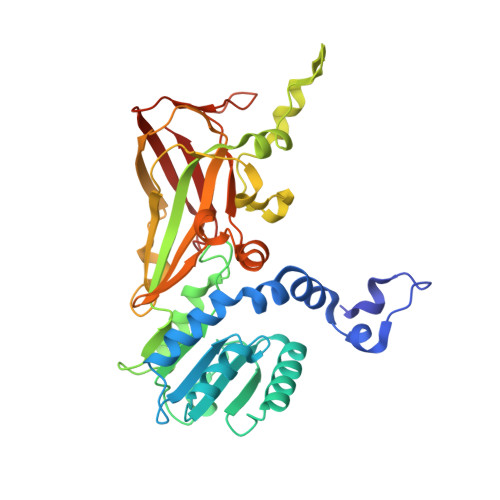
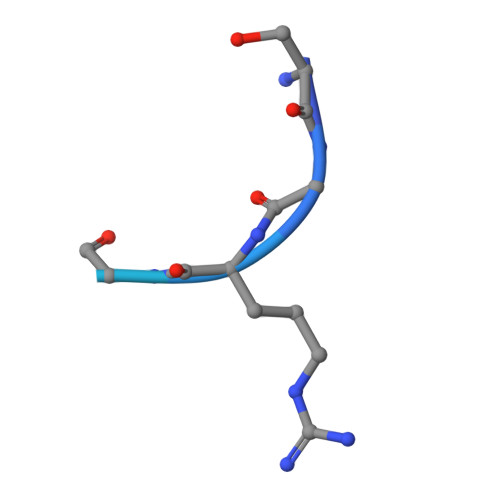
Structural determinants for the strict monomethylation activity by trypanosoma brucei protein arginine methyltransferase 7.
Wang, C., Zhu, Y., Caceres, T.B., Liu, L., Peng, J., Wang, J., Chen, J., Chen, X., Zhang, Z., Zuo, X., Gong, Q., Teng, M., Hevel, J.M., Wu, J., Shi, Y.(2014) Structure 22: 756-768
- PubMed: 24726341
- DOI: https://doi.org/10.1016/j.str.2014.03.003
- Primary Citation of Related Structures:
4M36, 4M37, 4M38 - PubMed Abstract:
Trypanosoma brucei protein arginine methyltransferase 7 (TbPRMT7) exclusively generates monomethylarginine (MMA), which directs biological consequences distinct from that of symmetric dimethylarginine (SDMA) and asymmetric dimethylarginine (ADMA). However, determinants controlling the strict monomethylation activity are unknown. We present the crystal structure of the TbPRMT7 active core in complex with S-adenosyl-L-homocysteine (AdoHcy) and a histone H4 peptide substrate. In the active site, residues E172, E181, and Q329 hydrogen bond the guanidino group of the target arginine and align the terminal guanidino nitrogen in a position suitable for nucleophilic attack on the methyl group of S-adenosyl-L-methionine (AdoMet). Structural comparisons and isothermal titration calorimetry data suggest that the TbPRMT7 active site is narrower than those of protein arginine dimethyltransferases, making it unsuitable to bind MMA in a manner that would support a second turnover, thus abolishing the production of SDMA and ADMA. Our results present the structural interpretations for the monomethylation activity of TbPRMT7.
- Hefei National Laboratory for Physical Sciences at Microscale and School of Life Sciences, University of Science and Technology of China, Anhui 230027, China.
Organizational Affiliation: