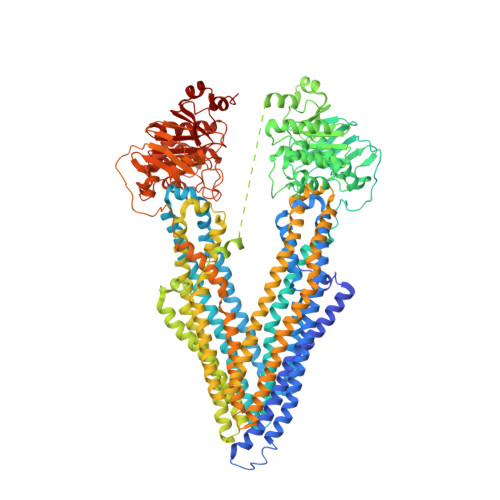
Refined structures of mouse P-glycoprotein.
Li, J., Jaimes, K.F., Aller, S.G.(2014) Protein Sci 23: 34-46
- PubMed: 24155053
- DOI: https://doi.org/10.1002/pro.2387
- Primary Citation of Related Structures:
4M1M, 4M2S, 4M2T - PubMed Abstract:
The recently determined C. elegans P-glycoprotein (Pgp) structure revealed significant deviations compared to the original mouse Pgp structure, which suggested possible misinterpretations in the latter model. To address this concern, we generated an experimental electron density map from single-wavelength anomalous dispersion phasing of an original mouse Pgp dataset to 3.8 Å resolution. The map exhibited significantly more detail compared to the original MAD map and revealed several regions of the structure that required de novo model building. The improved drug-free structure was refined to 3.8 Å resolution with a 9.4 and 8.1% decrease in R(work) and R(free), respectively, (R(work) = 21.2%, R(free) = 26.6%) and a significant improvement in protein geometry. The improved mouse Pgp model contains ∼95% of residues in the favorable Ramachandran region compared to only 57% for the original model. The registry of six transmembrane helices was corrected, revealing amino acid residues involved in drug binding that were previously unrecognized. Registry shifts (rotations and translations) for three transmembrane (TM)4 and TM5 and the addition of three N-terminal residues were necessary, and were validated with new mercury labeling and anomalous Fourier density. The corrected position of TM4, which forms the frame of a portal for drug entry, had backbone atoms shifted >6 Å from their original positions. The drug translocation pathway of mouse Pgp is 96% identical to human Pgp and is enriched in aromatic residues that likely play a collective role in allowing a high degree of polyspecific substrate recognition.
- Department of Pharmacology and Toxicology, Center for Structural Biology, University of Alabama at Birmingham, Birmingham, Alabama, 35205.
Organizational Affiliation: