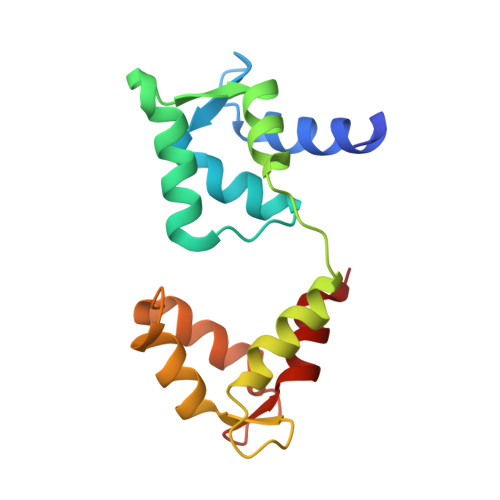
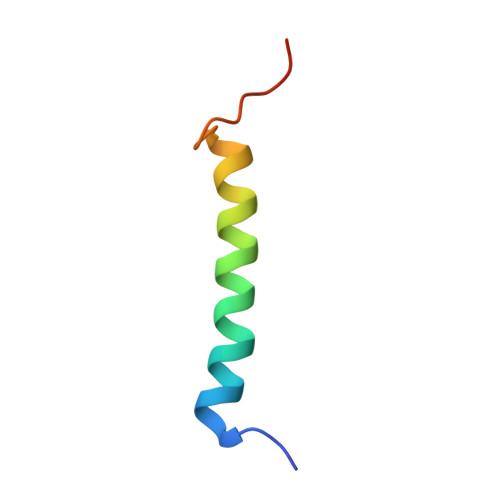
Functional and molecular features of the calmodulin-interacting protein IQCG required for haematopoiesis in zebrafish
Chen, L.T., Liang, W.X., Chen, S., Li, R.K., Tan, J.L., Xu, P.F., Luo, L.F., Wang, L., Yu, S.H., Meng, G., Li, K.K., Liu, T.X., Chen, Z., Chen, S.J.(2014) Nat Commun 5: 3811-3811
- PubMed: 24787902
- DOI: https://doi.org/10.1038/ncomms4811
- Primary Citation of Related Structures:
4LZX, 4M1L - PubMed Abstract:
We previously reported a fusion protein NUP98-IQCG in an acute leukaemia, which functions as an aberrant regulator of transcriptional expression, yet the structure and function of IQCG have not been characterized. Here we use zebrafish to investigate the role of iqcg in haematopoietic development, and find that the numbers of haematopoietic stem cells and multilineage-differentiated cells are reduced in iqcg-deficient embryos. Mechanistically, IQCG binds to calmodulin (CaM) and acts as a molecule upstream of CaM-dependent kinase IV (CaMKIV). Crystal structures of complexes between CaM and IQ domain of IQCG reveal dual CaM-binding footprints in this motif, and provide a structural basis for a higher CaM-IQCG affinity when deprived of calcium. The results collectively allow us to understand IQCG-mediated calcium signalling in haematopoiesis, and propose a model in which IQCG stores CaM at low cytoplasmic calcium concentrations, and releases CaM to activate CaMKIV when calcium level rises.
- 1] State Key Laboratory of Medical Genomics and Shanghai, Institute of Hematology, Rui Jin Hospital affiliated to Shanghai Jiao Tong University (SITU) School of Medicine, Shanghai, 200025, China [2] Institute of Health Sciences, Shanghai Institutes for Biological Sciences and Graduate School, Chinese Academy of Sciences and SITU School of Medicine, Shanghai 200025, China [3].
Organizational Affiliation: