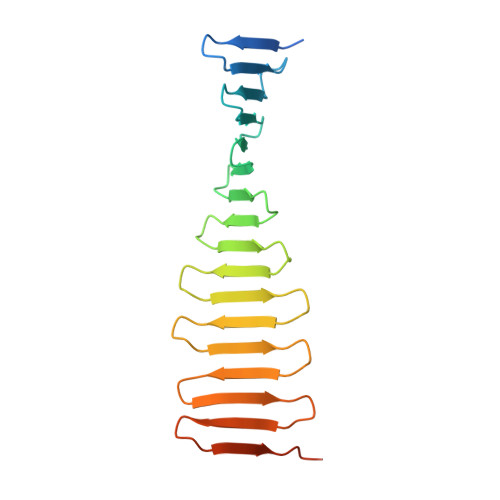
Structural analysis of the G-box domain of the microcephaly protein CPAP suggests a role in centriole architecture.
Hatzopoulos, G.N., Erat, M.C., Cutts, E., Rogala, K.B., Slater, L.M., Stansfeld, P.J., Vakonakis, I.(2013) Structure 21: 2069-2077
- PubMed: 24076405
- DOI: https://doi.org/10.1016/j.str.2013.08.019
- Primary Citation of Related Structures:
4LD1, 4LD3, 4LZF - PubMed Abstract:
Centrioles are evolutionarily conserved eukaryotic organelles composed of a protein scaffold surrounded by sets of microtubules organized with a 9-fold radial symmetry. CPAP, a centriolar protein essential for microtubule recruitment, features a C-terminal domain of unknown structure, the G-box. A missense mutation in the G-box reduces affinity for the centriolar shuttling protein STIL and causes primary microcephaly. Here, we characterize the molecular architecture of CPAP and determine the G-box structure alone and in complex with a STIL fragment. The G-box comprises a single elongated β sheet capable of forming supramolecular assemblies. Structural and biophysical studies highlight the conserved nature of the CPAP-STIL complex. We propose that CPAP acts as a horizontal "strut" that joins the centriolar scaffold with microtubules, whereas G-box domains form perpendicular connections.
- Department of Biochemistry, University of Oxford, South Parks Road, Oxford OX1 3QU, UK.
Organizational Affiliation: