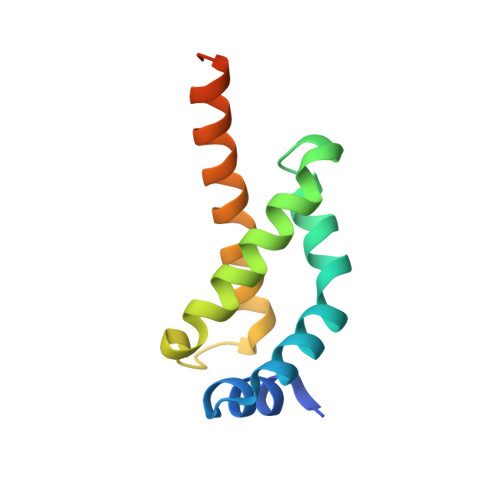
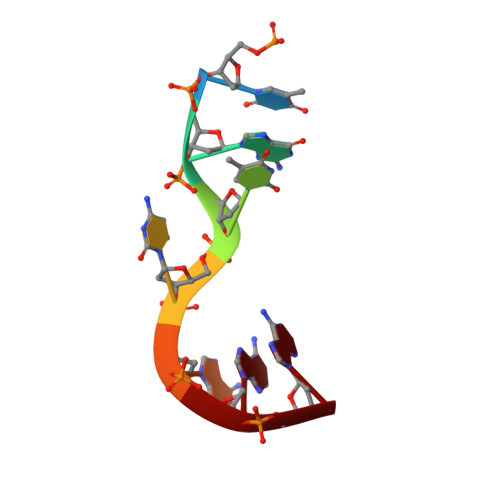Structural basis for -10 promoter element melting by environmentally induced sigma factors.
Campagne, S., Marsh, M.E., Capitani, G., Vorholt, J.A., Allain, F.H.(2014) Nat Struct Mol Biol 21: 269-276
- PubMed: 24531660
- DOI: https://doi.org/10.1038/nsmb.2777
- Primary Citation of Related Structures:
2MAO, 2MAP, 4LUP - PubMed Abstract:
Bacterial transcription is controlled by sigma factors, the RNA polymerase subunits that act as initiation factors. Although a single housekeeping sigma factor enables transcription from thousands of promoters, environmentally induced sigma factors redirect gene expression toward small regulons to carry out focused responses. Using structural and functional analyses, we determined the molecular basis of -10 promoter element recognition by Escherichia coli σ(E), which revealed an unprecedented way to achieve promoter melting. Group IV sigma factors induced strand separation at the -10 element by flipping out a single nucleotide from the nontemplate-strand DNA base stack. Unambiguous selection of this critical base was driven by a dynamic protein loop, which can be substituted to modify specificity of promoter recognition. This mechanism of promoter melting explains the increased promoter-selection stringency of environmentally induced sigma factors.
- 1] Institute of Microbiology, Eidgenössische Technische Hochschule (ETH) Zurich, Zurich, Switzerland. [2] Institute of Molecular Biology and Biophysics, Eidgenössische Technische Hochschule (ETH) Zurich, Zurich, Switzerland.
Organizational Affiliation: