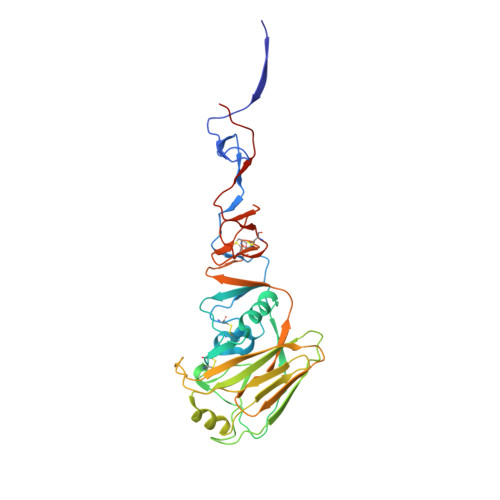
Structural Analysis of the Hemagglutinin from the Recent 2013 H7N9 Influenza Virus.
Yang, H., Carney, P.J., Chang, J.C., Villanueva, J.M., Stevens, J.(2013) J Virol 87: 12433-12446
- PubMed: 24027325
- DOI: https://doi.org/10.1128/JVI.01854-13
- Primary Citation of Related Structures:
4LN3, 4LN4, 4LN6, 4LN8 - PubMed Abstract:
In March 2013, the Chinese Center for Disease Control and Prevention reported human infections with an H7N9 influenza virus, and by 20 July 2013, the numbers of laboratory-confirmed cases had climbed to 134, including 43 fatalities and 127 hospitalizations. The newly emerging H7N9 viruses constitute an obvious public health concern because of the apparent severity of this outbreak. Here we focus on the hemagglutinins (HAs) of these viruses and assess their receptor binding phenotype in relation to previous HAs studied. Glycan microarray and kinetic analyses of recombinant A(H7N9) HAs were performed to compare the receptor binding profile of wild-type receptor binding site variants at position 217, a residue analogous to one of two positions known to switch avian to human receptor preference in H2N2 and H3N2 viruses. Two recombinant A(H7N9) HAs were structurally characterized, and a mutational study of the receptor binding site was performed to analyze important residues that can affect receptor preference and affinity. Results highlight a weak human receptor preference of the H7N9 HAs, suggesting that these viruses require further adaptation in order to adapt fully to humans.
- Influenza Division, National Center for Immunization and Respiratory Diseases, Centers for Disease Control and Prevention, Atlanta, Georgia, USA.
Organizational Affiliation: