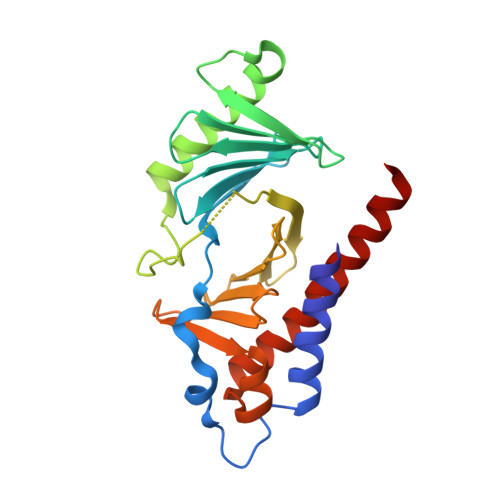
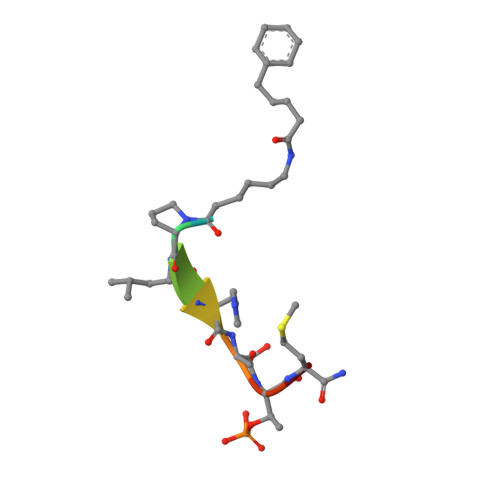
Exploring the binding nature of pyrrolidine pocket-dependent interactions in the polo-box domain of polo-like kinase 1
Murugan, R.N., Ahn, M., Lee, W.C., Kim, H.Y., Song, J.H., Cheong, C., Hwang, E., Seo, J.H., Shin, S.Y., Choi, S.H., Park, J.E., Bang, J.K.(2013) PLoS One 8: e80043-e80043
- PubMed: 24223211
- DOI: https://doi.org/10.1371/journal.pone.0080043
- Primary Citation of Related Structures:
4LKL, 4LKM - PubMed Abstract:
Over the years, a great deal of effort has been focused on the design and synthesis of potent, linear peptide inhibitors targeting the polo-like kinase 1 (Plk1), which is critically involved in multiple mitotic processes and has been established as an adverse prognostic marker for tumor patients. Plk1 localizes to its intracellular anchoring sites via its polo-box domain, and inhibiting the Plk1 polo-box domain has been considered as an approach to circumvent the specificity problems associated with inhibiting the conserved adenosine triphosphate-binding pocket. The polo-box domain consists of two different binding regions, such as the unique, broader pyrrolidine-binding pocket and the conserved, narrow, Tyr-rich hydrophobic channel, among the three Plk polo-box domains (Plks 1-3), respectively. Therefore, the studies that provide insights into the binding nature of the unique, broader pyrrolidine-binding pocket might lead to the development of selective Plk1-inhibitory compounds. In an attempt to retain the monospecificity by targeting the unique, broader pyrrolidine-binding pocket, here, for the first time, a systematic approach was undertaken to examine the structure-activity relationship of N-terminal-truncated PLHSpTM derivatives, to apply a site-directed ligand approach using bulky aromatic and non-aromatic systems, and to characterize the binding nature of these analogues using X-ray crystallographic studies. We have identified a new mode of binding interactions, having improved binding affinity and retaining the Plk1 polo-box domain specificity, at the pyrrolidine-binding pocket. Furthermore, our data revealed that the pyrrolidine-binding pocket was very specific to recognize a short and bulky hydrophobic ligand like adamantane, whereas the Tyr-rich hydrophobic channel was specific with lengthy and small hydrophobic groups. The progress made using our site-directed ligands validated this approach to specifically direct the ligand into the unique pyrrolidine-binding region, and it extends the applicability of the strategy for discovering selective protein-protein interaction inhibitors.
- Division of Magnetic Resonance, Korea Basic Science Institute, Ochang, Chung-Buk, Republic of Korea.
Organizational Affiliation: