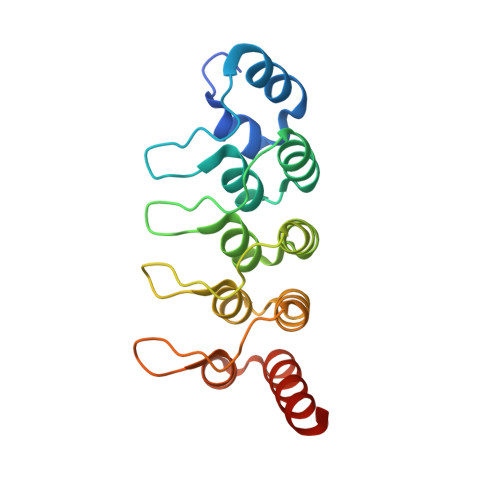
Ankyrin Repeats of ANKRA2 Recognize a PxLPxL Motif on the 3M Syndrome Protein CCDC8.
Nie, J., Xu, C., Jin, J., Aka, J.A., Tempel, W., Nguyen, V., You, L., Weist, R., Min, J., Pawson, T., Yang, X.J.(2015) Structure 23: 700-712
- PubMed: 25752541
- DOI: https://doi.org/10.1016/j.str.2015.02.001
- Primary Citation of Related Structures:
4LG6 - PubMed Abstract:
Peptide motifs are often used for protein-protein interactions. We have recently demonstrated that ankyrin repeats of ANKRA2 and the paralogous bare lymphocyte syndrome transcription factor RFXANK recognize PxLPxL/I motifs shared by megalin, three histone deacetylases, and RFX5. We show here that that CCDC8 is a major partner of ANKRA2 but not RFXANK in cells. The CCDC8 gene is mutated in 3M syndrome, a short-stature disorder with additional facial and skeletal abnormalities. Two other genes mutated in this syndrome encode CUL7 and OBSL1. While CUL7 is a ubiquitin ligase and OBSL1 associates with the cytoskeleton, little is known about CCDC8. Binding and structural analyses reveal that the ankyrin repeats of ANKRA2 recognize a PxLPxL motif at the C-terminal region of CCDC8. The N-terminal part interacts with OBSL1 to form a CUL7 ligase complex. These results link ANKRA2 unexpectedly to 3M syndrome and suggest novel regulatory mechanisms for histone deacetylases and RFX7.
- The Rosalind & Morris Goodman Cancer Research Center, Department of Medicine, McGill University, Montréal, QC H3A 1A3, Canada; Department of Breast Cancer, The Third Affiliated Hospital, Kunming Medical University, Yunnan 650118, China.
Organizational Affiliation: