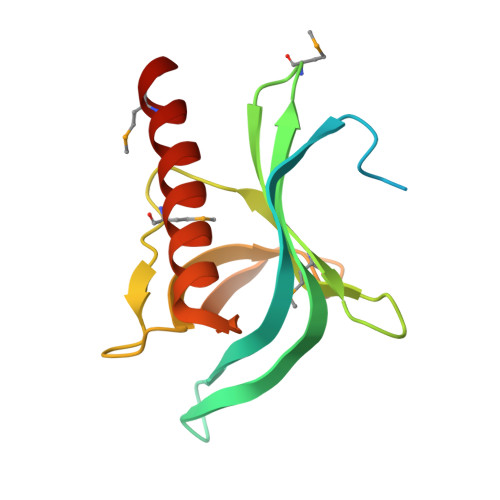
Crystal Structure of the RanBD1 fourth domain of E3 SUMO-protein ligase RanBP2.
Vorobiev, S., Su, M., Seetharaman, J., Mao, L., Xiao, R., Maglaqui, M., Kogan, S., Wang, H., Everett, J.K., Acton, T.B., Montelione, G.T., Hunt, J.F., Tong, L.To be published.