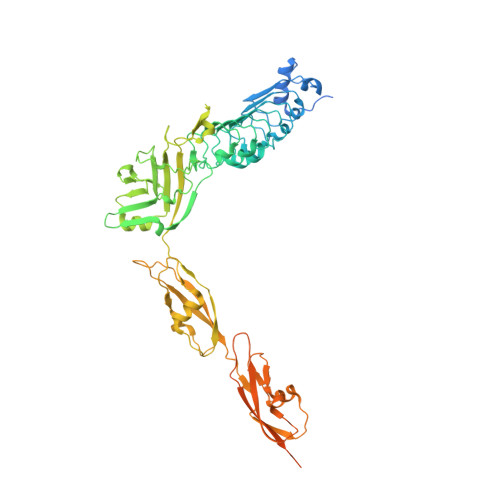
Structure of Internalin InlK from the Human Pathogen Listeria monocytogenes.
Neves, D., Job, V., Dortet, L., Cossart, P., Dessen, A.(2013) J Mol Biology 425: 4520-4529
- PubMed: 23958637
- DOI: https://doi.org/10.1016/j.jmb.2013.08.010
- Primary Citation of Related Structures:
4L3A, 4L3F - PubMed Abstract:
Listeria monocytogenes is a human pathogen that employs a wide variety of virulence factors in order to adhere to, invade, and replicate within target cells. Internalins play key roles in processes ranging from adhesion to receptor recognition and are thus essential for infection. Recently, InlK, a surface-associated internalin, was shown to be involved in Listeria's ability to escape from autophagy by recruitment of the major vault protein (MVP) to the bacterial surface. Here, we report the structure of InlK, which harbors four domains arranged in the shape of a "bent arm". The structure supports a role for the "elbow" of InlK in partner recognition, as well as of two Ig-like pedestals intercalated by hinge regions in the projection of InlK away from the surface of the bacterium. The unusual fold and flexibility of InlK could be essential for MVP binding and concealment from recognition by molecules involved in the autophagic process.
- Brazilian National Laboratory for Biosciences (LNBio), CNPEM, Campinas, São Paulo, Brazil.
Organizational Affiliation: