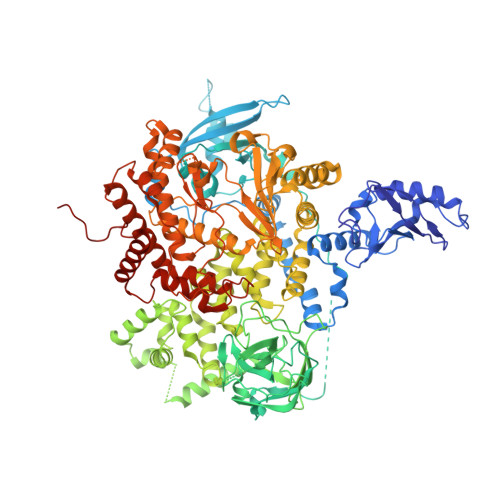
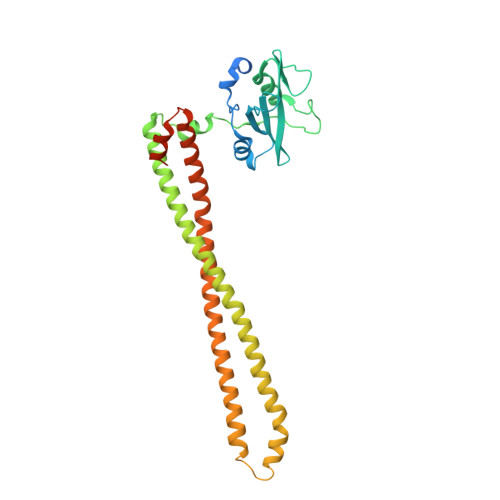
Crystal Structures of PI3K alpha Complexed with PI103 and Its Derivatives: New Directions for Inhibitors Design.
Zhao, Y., Zhang, X., Chen, Y., Lu, S., Peng, Y., Wang, X., Guo, C., Zhou, A., Zhang, J., Luo, Y., Shen, Q., Ding, J., Meng, L., Zhang, J.(2014) ACS Med Chem Lett 5: 138-142
- PubMed: 24900786
- DOI: https://doi.org/10.1021/ml400378e
- Primary Citation of Related Structures:
4L1B, 4L23, 4L2Y - PubMed Abstract:
The phosphatidylinositol 3-kinase (PI3K) signaling pathway plays important roles in cell proliferation, growth, and survival. Hyperactivated PI3K is frequently found in a wide variety of human cancers, validating it as a promising target for cancer therapy. We determined the crystal structure of the human PI3Kα-PI103 complex to unravel molecular interactions. Based on the structure, substitution at the R1 position of the phenol portion of PI103 was demonstrated to improve binding affinity via forming a new H-bond with Lys802 at the bottom of the ATP catalytic site. Interestingly, the crystal structure of the PI3Kα-9d complex revealed that the flexibility of Lys802 can also induce additional space at the catalytic site for further modification. Thus, these crystal structures provide a molecular basis for the strong and specific interactions and demonstrate the important role of Lys802 in the design of novel PI3Kα inhibitors.
- Department of Pathophysiology, Key Laboratory of Cell Differentiation and Apoptosis of Chinese Ministry of Education, Shanghai JiaoTong University, School of Medicine , Shanghai 200025, China.
Organizational Affiliation: