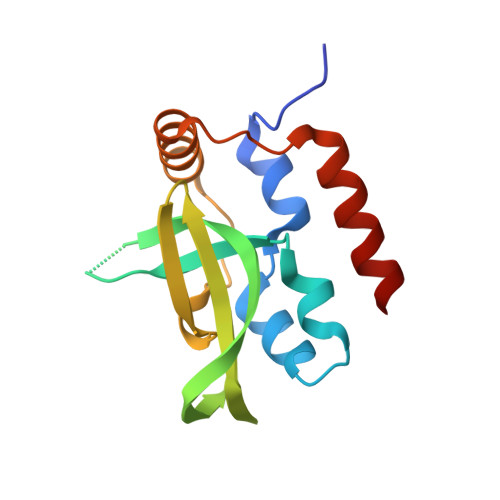
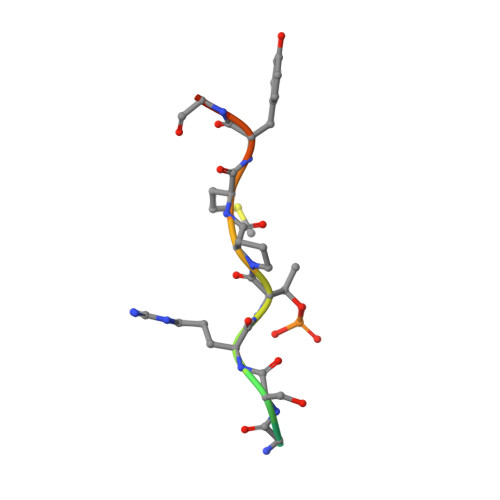
Structural basis for Spt5-mediated recruitment of the Paf1 complex to chromatin.
Wier, A.D., Mayekar, M.K., Heroux, A., Arndt, K.M., Vandemark, A.P.(2013) Proc Natl Acad Sci U S A 110: 17290-17295
- PubMed: 24101474
- DOI: https://doi.org/10.1073/pnas.1314754110
- Primary Citation of Related Structures:
4L1P, 4L1U - PubMed Abstract:
Polymerase associated factor 1 complex (Paf1C) broadly influences gene expression by regulating chromatin structure and the recruitment of RNA-processing factors during transcription elongation. The Plus3 domain of the Rtf1 subunit mediates Paf1C recruitment to genes by binding a repeating domain within the elongation factor Spt5 (suppressor of Ty). Here we provide a molecular description of this interaction by reporting the structure of human Rtf1 Plus3 in complex with a phosphorylated Spt5 repeat. We find that Spt5 binding is mediated by an extended surface containing phosphothreonine recognition and hydrophobic interfaces that interact with residues outside the Spt5 motif. Changes within these interfaces diminish binding of Spt5 in vitro and chromatin localization of Rtf1 in vivo. The structure reveals the basis for recognition of the repeat motif of Spt5, a key player in the recruitment of gene regulatory factors to RNA polymerase II.
- Department of Biological Sciences, University of Pittsburgh, Pittsburgh, PA 15260.
Organizational Affiliation: