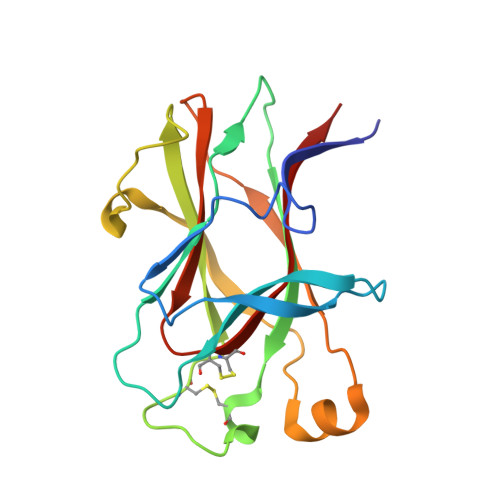
Distinctive Structure of the EphA3/Ephrin-A5 Complex Reveals a Dual Mode of Eph Receptor Interaction for Ephrin-A5.
Forse, G.J., Uson, M.L., Nasertorabi, F., Kolatkar, A., Lamberto, I., Pasquale, E.B., Kuhn, P.(2015) PLoS One 10: e0127081-e0127081
- PubMed: 25993310
- DOI: https://doi.org/10.1371/journal.pone.0127081
- Primary Citation of Related Structures:
4L0P - PubMed Abstract:
The Eph receptor tyrosine kinase/ephrin ligand system regulates a wide spectrum of physiological processes, while its dysregulation has been implicated in cancer progression. The human EphA3 receptor is widely upregulated in the tumor microenvironment and is highly expressed in some types of cancer cells. Furthermore, EphA3 is among the most highly mutated genes in lung cancer and it is also frequently mutated in other cancers. We report the structure of the ligand-binding domain of the EphA3 receptor in complex with its preferred ligand, ephrin-A5. The structure of the complex reveals a pronounced tilt of the ephrin-A5 ligand compared to its orientation when bound to the EphA2 and EphB2 receptors and similar to its orientation when bound to EphA4. This tilt brings an additional area of ephrin-A5 into contact with regions of EphA3 outside the ephrin-binding pocket thereby enlarging the size of the interface, which is consistent with the high binding affinity of ephrin-A5 for EphA3. This large variation in the tilt of ephrin-A5 bound to different Eph receptors has not been previously observed for other ephrins.
- Dornsife College of Letters, Arts and Sciences, University of Southern California, 3430 S. Vermont Ave., Suite 105 (110), MC3301, Los Angeles, CA, 90089-3301, United States of America.
Organizational Affiliation: