Northeast Structural Genomics Consortium Target OR342
Kuzin, A., Lew, S., Rajagopalan, S., Seetharaman, J., Mao, L., Xiao, R., Lee, D., Raja, S., Everett, J.K., Acton, T.B., Baker, D., Montelione, G.T., Tong, L., Hunt, J.F.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Designed Protein OR342 | 167 | synthetic construct | Mutation(s): 0 | 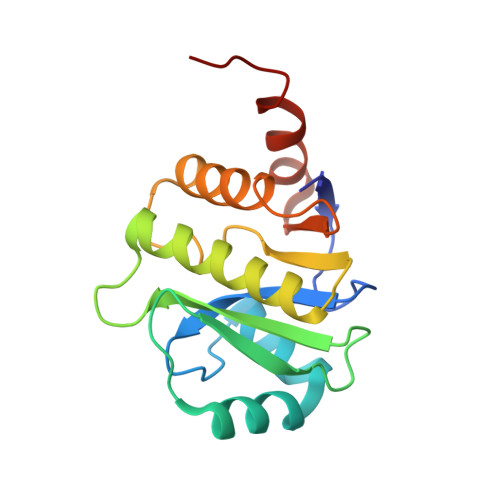 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| PO4 Query on PO4 | C [auth A], D [auth A], E [auth B], F [auth B] | PHOSPHATE ION O4 P NBIIXXVUZAFLBC-UHFFFAOYSA-K |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 30.054 | α = 90 |
| b = 76.401 | β = 90 |
| c = 132.584 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| PDB_EXTRACT | data extraction |
| HKL-2000 | data collection |
| DENZO | data reduction |
| SCALEPACK | data scaling |
| BALBES | phasing |