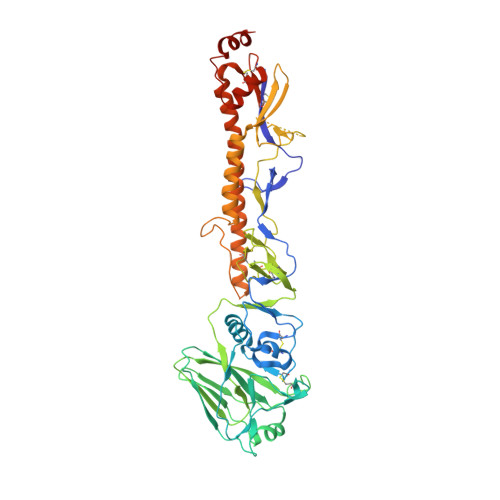
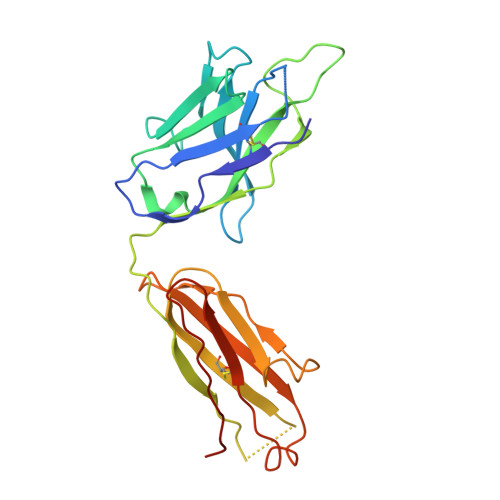
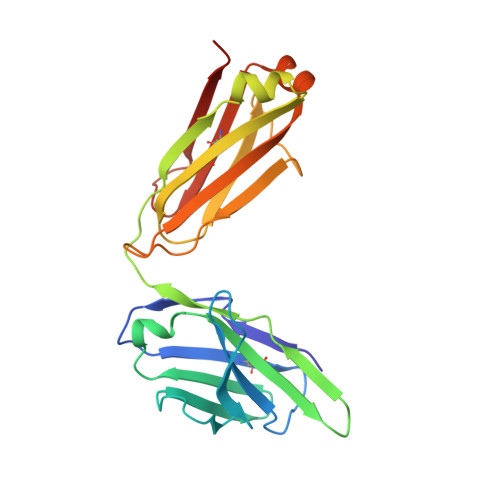
A Novel In vivo Human Plasmablast Enrichment Technique Allows Rapid Identification of Therapeutic Anti-Influenza A Antibodies
Nakamura, G., Chai, N., Park, S., Chiang, N., Lin, Z., Chiu, H., Fong, R., Yan, D., Kim, J., Zhang, J., Lee, W.P., Estevez, A., Coons, M., Xu, M., Lupardus, P., Balazs, M., Swem, L.R.(2013) Cell Host Microbe 14: 93-103
- PubMed: 23870317
- DOI: https://doi.org/10.1016/j.chom.2013.06.004
- Primary Citation of Related Structures:
4KVN - PubMed Abstract:
Recent advances enabling the cloning of human immunoglobulin G genes have proven effective for discovering monoclonal antibodies with therapeutic potential. However, these antibody-discovery methods are often arduous and identify only a few candidates from numerous antibody-secreting plasma cells or plasmablasts. We describe an in vivo enrichment technique that identifies broadly neutralizing human antibodies with high frequency. For this technique, human peripheral blood mononuclear cells from vaccinated donors are activated and enriched in an antigen-specific manner for the production of numerous antigen-specific plasmablasts. Using this technology, we identified four broadly neutralizing influenza A antibodies by screening only 840 human antibodies. Two of these antibodies neutralize every influenza A human isolate tested and perform better than the current anti-influenza A therapeutic, oseltamivir, in treating severe influenza infection in mice and ferrets. Furthermore, these antibodies elicit robust in vivo synergism when combined with oseltamivir, thus highlighting treatment strategies that could benefit influenza-infected patients.
- Antibody Engineering Department, Genentech, South San Francisco, CA 94080, USA.
Organizational Affiliation: