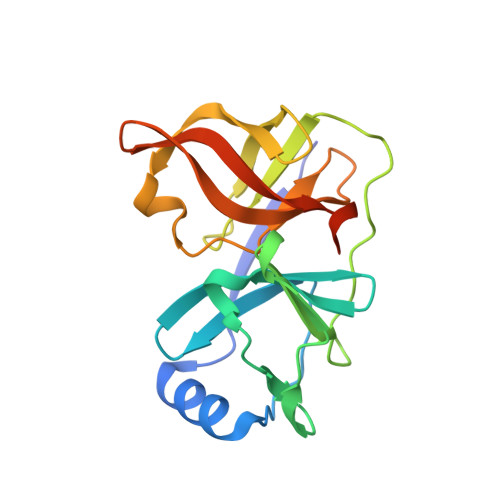
Discovery of Danoprevir (ITMN-191/R7227), a Highly Selective and Potent Inhibitor of Hepatitis C Virus (HCV) NS3/4A Protease.
Jiang, Y., Andrews, S.W., Condroski, K.R., Buckman, B., Serebryany, V., Wenglowsky, S., Kennedy, A.L., Madduru, M.R., Wang, B., Lyon, M., Doherty, G.A., Woodard, B.T., Lemieux, C., Do, M.G., Zhang, H., Ballard, J., Vigers, G., Brandhuber, B.J., Stengel, P., Josey, J.A., Beigelman, L., Blatt, L., Seiwert, S.D.(2014) J Med Chem 57: 1753-1769
- PubMed: 23672640
- DOI: https://doi.org/10.1021/jm400164c
- Primary Citation of Related Structures:
4KTC - PubMed Abstract:
HCV serine protease NS3 represents an attractive drug target because it is not only essential for viral replication but also implicated in the viral evasion of the host immune response pathway through direct cleavage of key proteins in the human innate immune system. Through structure-based drug design and optimization, macrocyclic peptidomimetic molecules bearing both a lipophilic P2 isoindoline carbamate and a P1/P1' acylsulfonamide/acylsulfamide carboxylic acid bioisostere were prepared that possessed subnanomolar potency against the NS3 protease in a subgenomic replicon-based cellular assay (Huh-7). Danoprevir (compound 49) was selected as the clinical development candidate for its favorable potency profile across multiple HCV genotypes and key mutant strains and for its good in vitro ADME profiles and in vivo target tissue (liver) exposures across multiple animal species. X-ray crystallographic studies elucidated several key features in the binding of danoprevir to HCV NS3 protease and proved invaluable to our iterative structure-based design strategy.
- Array BioPharma , 3200 Walnut Street, Boulder, Colorado 80301, United States.
Organizational Affiliation: