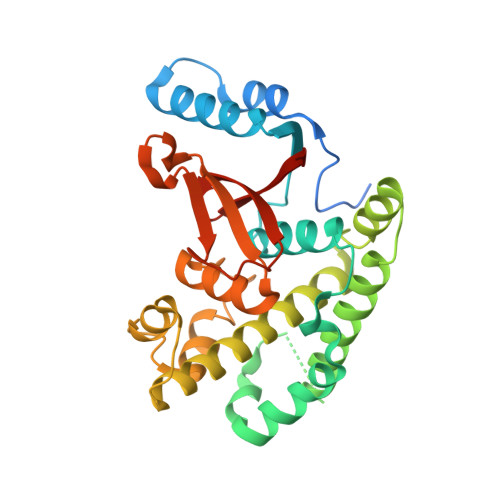
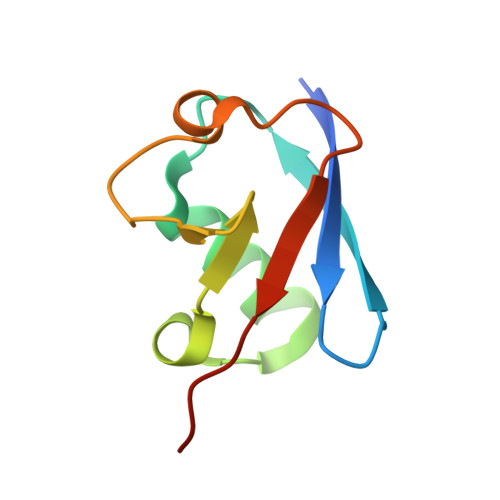
The linear ubiquitin-specific deubiquitinase gumby regulates angiogenesis.
Rivkin, E., Almeida, S.M., Ceccarelli, D.F., Juang, Y.C., MacLean, T.A., Srikumar, T., Huang, H., Dunham, W.H., Fukumura, R., Xie, G., Gondo, Y., Raught, B., Gingras, A.C., Sicheri, F., Cordes, S.P.(2013) Nature 498: 318-324
- PubMed: 23708998
- DOI: https://doi.org/10.1038/nature12296
- Primary Citation of Related Structures:
4KSJ, 4KSK, 4KSL - PubMed Abstract:
A complex interaction of signalling events, including the Wnt pathway, regulates sprouting of blood vessels from pre-existing vasculature during angiogenesis. Here we show that two distinct mutations in the (uro)chordate-specific gumby (also called Fam105b) gene cause an embryonic angiogenic phenotype in gumby mice. Gumby interacts with disheveled 2 (DVL2), is expressed in canonical Wnt-responsive endothelial cells and encodes an ovarian tumour domain class of deubiquitinase that specifically cleaves linear ubiquitin linkages. A crystal structure of gumby in complex with linear diubiquitin reveals how the identified mutations adversely affect substrate binding and catalytic function in line with the severity of their angiogenic phenotypes. Gumby interacts with HOIP (also called RNF31), a key component of the linear ubiquitin assembly complex, and decreases linear ubiquitination and activation of NF-κB-dependent transcription. This work provides support for the biological importance of linear (de)ubiquitination in angiogenesis, craniofacial and neural development and in modulating Wnt signalling.
- Samuel Lunenfeld Research Institute, Mt Sinai Hospital, 600 University Avenue, Toronto, Ontario M5G 1X5, Canada.
Organizational Affiliation: