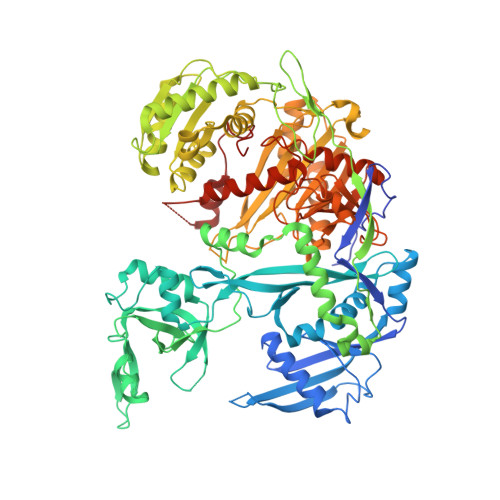
The making of a slicer: activation of human argonaute-1.
Faehnle, C.R., Elkayam, E., Haase, A.D., Hannon, G.J., Joshua-Tor, L.(2013) Cell Rep 3: 1901-1909
- PubMed: 23746446
- DOI: https://doi.org/10.1016/j.celrep.2013.05.033
- Primary Citation of Related Structures:
4KRE, 4KRF - PubMed Abstract:
Argonautes are the central protein component in small RNA silencing pathways. Of the four human Argonautes (hAgo1-hAgo4) only hAgo2 is an active slicer. We determined the structure of hAgo1 bound to endogenous copurified RNAs to 1.75 Å resolution and hAgo1 loaded with let-7 microRNA to 2.1 Å. Both structures are strikingly similar to the structures of hAgo2. A conserved catalytic tetrad within the PIWI domain of hAgo2 is required for its slicing activity. Completion of the tetrad, combined with a mutation on a loop adjacent to the active site of hAgo1, results in slicer activity that is substantially enhanced by swapping in the N domain of hAgo2. hAgo3, with an intact tetrad, becomes an active slicer by swapping the N domain of hAgo2 without additional mutations. Intriguingly, the elements that make Argonaute an active slicer involve a sophisticated interplay between the active site and more distant regions of the enzyme.
- W.M. Keck Structural Biology Laboratory, Cold Spring Harbor Laboratory, 1 Bungtown Road, Cold Spring Harbor, NY 11724, USA.
Organizational Affiliation: