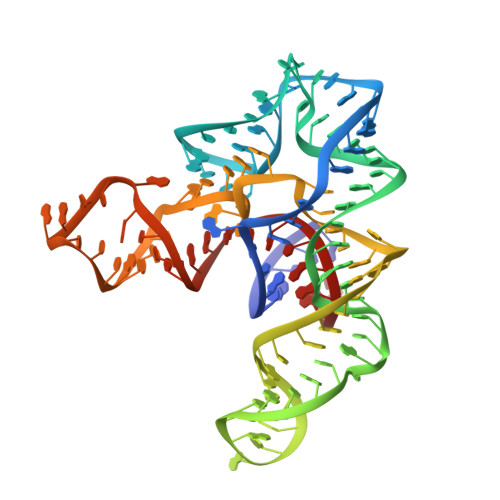
SAM recognition and conformational switching mechanism in the Bacillus subtilis yitJ S box/SAM-I riboswitch
Lu, C., Ding, F., Chowdhury, A., Pradhan, V., Tomsic, J., Holmes, W.M., Henkin, T.M., Ke, A.(2010) J Mol Biology 404: 803-818
- PubMed: 20951706
- DOI: https://doi.org/10.1016/j.jmb.2010.09.059
- Primary Citation of Related Structures:
4KQY - PubMed Abstract:
S-box (SAM-I) riboswitches are a widespread class of riboswitches involved in the regulation of sulfur metabolism in Gram-positive bacteria. We report here the 3.0-Å crystal structure of the aptamer domain of the Bacillus subtilis yitJ S-box (SAM-I) riboswitch bound to S-adenosyl-L-methionine (SAM). The RNA folds into two sets of helical stacks spatially arranged by tertiary interactions including a K-turn and a pseudoknot at a four-way junction. The tertiary structure is further stabilized by metal coordination, extensive ribose zipper interactions, and SAM-mediated tertiary interactions. Despite structural differences in the peripheral regions, the SAM-binding core of the B. subtilis yitJ riboswitch is virtually superimposable with the previously determined Thermoanaerobacter tengcongensis yitJ riboswitch structure, suggesting that a highly conserved ligand-recognition mechanism is utilized by all S-box riboswitches. SHAPE (selective 2'-hydroxyl acylation analyzed by primer extension) chemical probing analysis further revealed that the alternative base-pairing element in the expression platform controls the conformational switching process. In the absence of SAM, the apo yitJ aptamer domain folds predominantly into a pre-binding conformation that resembles, but is not identical with, the SAM-bound state. We propose that SAM enters the ligand-binding site through the "J1/2-J3/4" gate and "locks" down the SAM-bound conformation through an induced-fit mechanism. Temperature-dependent SHAPE revealed that the tertiary interaction-stabilized SAM-binding core is extremely stable, likely due to the cooperative RNA folding behavior. Mutational studies revealed that certain modifications in the SAM-binding region result in loss of SAM binding and constitutive termination, which suggests that these mutations lock the RNA into a form that resembles the SAM-bound form in the absence of SAM.
- Department of Molecular Biology and Genetics, 251 Biotechnology Building, Cornell University, Ithaca, NY 14853, USA.
Organizational Affiliation: