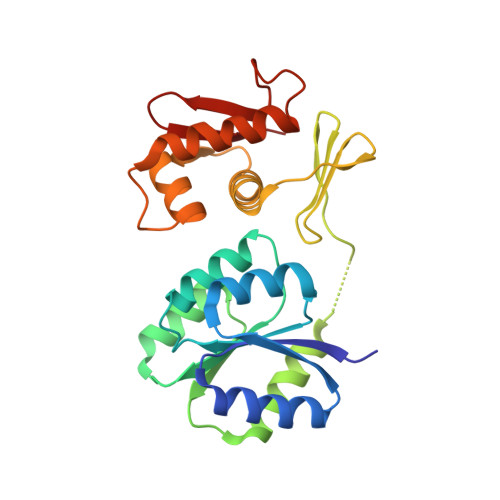
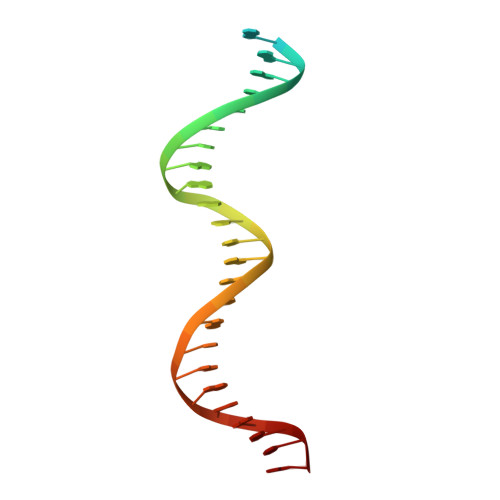
An asymmetric heterodomain interface stabilizes a response regulator-DNA complex.
Narayanan, A., Kumar, S., Evrard, A.N., Paul, L.N., Yernool, D.A.(2014) Nat Commun 5: 3282-3282
- PubMed: 24526190
- DOI: https://doi.org/10.1038/ncomms4282
- Primary Citation of Related Structures:
4KFC, 4KNY, 4L85 - PubMed Abstract:
Two-component signal transduction systems consist of pairs of histidine kinases and response regulators, which mediate adaptive responses to environmental cues. Most activated response regulators regulate transcription by binding tightly to promoter DNA via a phosphorylation-triggered inactive-to-active transition. The molecular basis for formation of stable response regulator-DNA complexes that precede the assembly of RNA polymerases is unclear. Here, we present structures of DNA complexed with the response regulator KdpE, a member of the OmpR/PhoB family. The distinctively asymmetric complex in an active-like conformation reveals a unique intramolecular interface between the receiver domain (RD) and the DNA-binding domain (DBD) of only one of the two response regulators in the complex. Structure-function studies show that this RD-DBD interface is necessary to form stable complexes that support gene expression. The conservation of sequence and structure suggests that these findings extend to a large group of response regulators that act as transcription factors.
- 1] Department of Biological Sciences, Purdue University, 915 West State Street, West Lafayette, Indiana 47907, USA [2].
Organizational Affiliation: