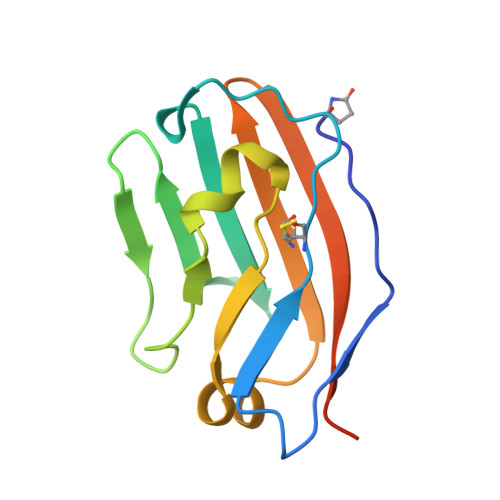
Engineered SIRP alpha variants as immunotherapeutic adjuvants to anticancer antibodies.
Weiskopf, K., Ring, A.M., Ho, C.C., Volkmer, J.P., Levin, A.M., Volkmer, A.K., Ozkan, E., Fernhoff, N.B., van de Rijn, M., Weissman, I.L., Garcia, K.C.(2013) Science 341: 88-91
- PubMed: 23722425
- DOI: https://doi.org/10.1126/science.1238856
- Primary Citation of Related Structures:
4KJY - PubMed Abstract:
CD47 is an antiphagocytic signal that cancer cells employ to inhibit macrophage-mediated destruction. Here, we modified the binding domain of human SIRPα, the receptor for CD47, for use as a CD47 antagonist. We engineered high-affinity SIRPα variants with about a 50,000-fold increased affinity for human CD47 relative to wild-type SIRPα. As high-affinity SIRPα monomers, they potently antagonized CD47 on cancer cells but did not induce macrophage phagocytosis on their own. Instead, they exhibited remarkable synergy with all tumor-specific monoclonal antibodies tested by increasing phagocytosis in vitro and enhancing antitumor responses in vivo. This "one-two punch" directs immune responses against tumor cells while lowering the threshold for macrophage activation, thereby providing a universal method for augmenting the efficacy of therapeutic anticancer antibodies.
- Institute for Stem Cell Biology and Regenerative Medicine, Stanford University School of Medicine, Stanford, CA 94305, USA.
Organizational Affiliation: