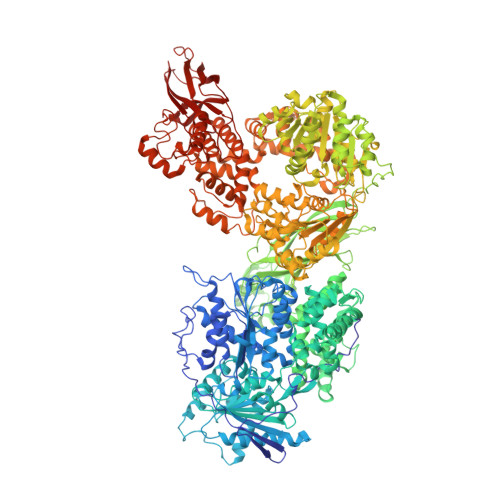
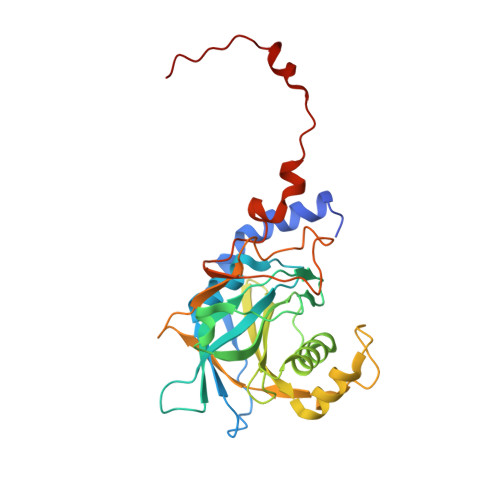
Inhibition of RNA helicase Brr2 by the C-terminal tail of the spliceosomal protein Prp8.
Mozaffari-Jovin, S., Wandersleben, T., Santos, K.F., Will, C.L., Luhrmann, R., Wahl, M.C.(2013) Science 341: 80-84
- PubMed: 23704370
- DOI: https://doi.org/10.1126/science.1237515
- Primary Citation of Related Structures:
4KIT - PubMed Abstract:
The Ski2-like RNA helicase Brr2 is a core component of the spliceosome that must be tightly regulated to ensure correct timing of spliceosome activation. Little is known about mechanisms of regulation of Ski2-like helicases by protein cofactors. Here we show by crystal structure and biochemical analyses that the Prp8 protein, a major regulator of the spliceosome, can insert its C-terminal tail into Brr2's RNA-binding tunnel, thereby intermittently blocking Brr2's RNA-binding, adenosine triphosphatase, and U4/U6 unwinding activities. Inefficient Brr2 repression is the only recognizable phenotype associated with certain retinitis pigmentosa-linked Prp8 mutations that map to its C-terminal tail. Our data show how a Ski2-like RNA helicase can be reversibly inhibited by a protein cofactor that directly competes with RNA substrate binding.
- Department of Cellular Biochemistry, Max Planck Institute for Biophysical Chemistry, Göttingen, Germany.
Organizational Affiliation: