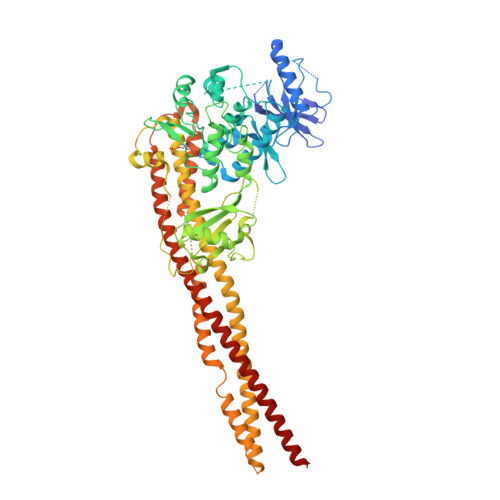
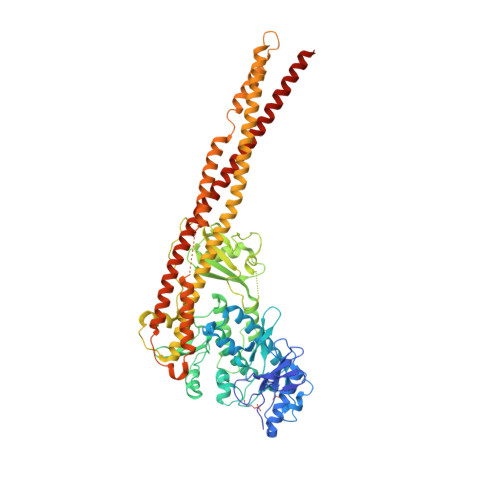
Crystal Structure of a Human I kappa B Kinase beta Asymmetric Dimer.
Liu, S., Misquitta, Y.R., Olland, A., Johnson, M.A., Kelleher, K.S., Kriz, R., Lin, L.L., Stahl, M., Mosyak, L.(2013) J Biological Chem 288: 22758-22767
- PubMed: 23792959
- DOI: https://doi.org/10.1074/jbc.M113.482596
- Primary Citation of Related Structures:
4KIK - PubMed Abstract:
Phosphorylation of inhibitor of nuclear transcription factor κB (IκB) by IκB kinase (IKK) triggers the degradation of IκB and migration of cytoplasmic κB to the nucleus where it promotes the transcription of its target genes. Activation of IKK is achieved by phosphorylation of its main subunit, IKKβ, at the activation loop sites. Here, we report the 2.8 Å resolution crystal structure of human IKKβ (hIKKβ), which is partially phosphorylated and bound to the staurosporine analog K252a. The hIKKβ protomer adopts a trimodular structure that closely resembles that from Xenopus laevis (xIKKβ): an N-terminal kinase domain (KD), a central ubiquitin-like domain (ULD), and a C-terminal scaffold/dimerization domain (SDD). Although hIKKβ and xIKKβ utilize a similar dimerization mode, their overall geometries are distinct. In contrast to the structure resembling closed shears reported previously for xIKKβ, hIKKβ exists as an open asymmetric dimer in which the two KDs are further apart, with one in an active and the other in an inactive conformation. Dimer interactions are limited to the C-terminal six-helix bundle that acts as a hinge between the two subunits. The observed domain movements in the structures of IKKβ may represent trans-phosphorylation steps that accompany IKKβ activation.
- Structural Biology and Biophysics Group, Pfizer Worldwide Research, Groton, Connecticut 06340, USA. shenping.liu@pfizer.com
Organizational Affiliation: