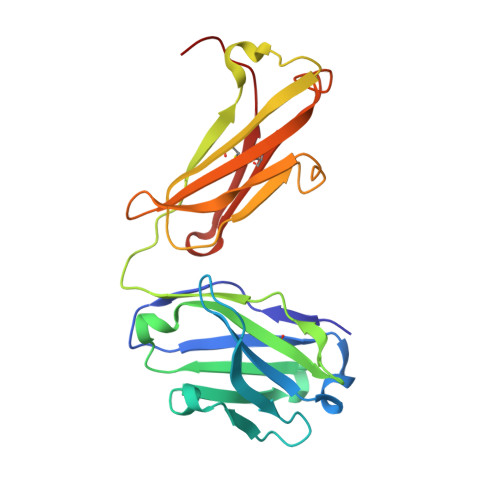
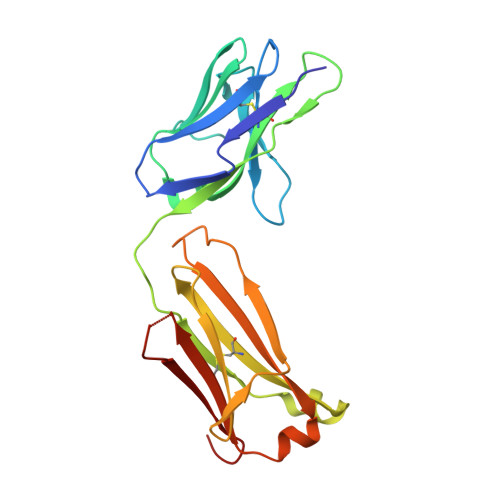
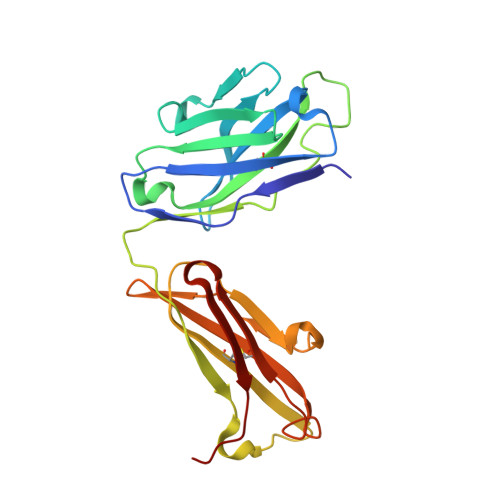
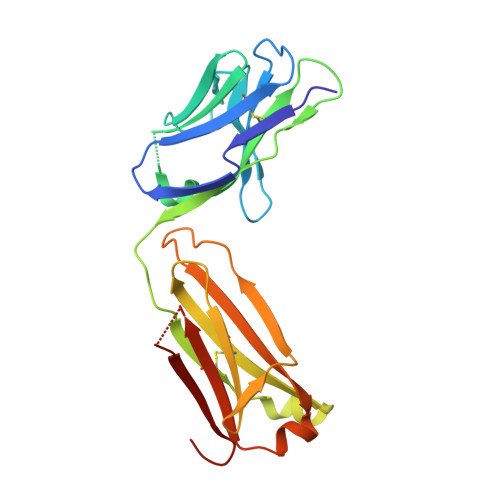
Structure of the factor VIII C2 domain in a ternary complex with 2 inhibitor antibodies reveals classical and nonclassical epitopes.
Walter, J.D., Werther, R.A., Brison, C.M., Cragerud, R.K., Healey, J.F., Meeks, S.L., Lollar, P., Spiegel, P.C.(2013) Blood 122: 4270-4278
- PubMed: 24085769
- DOI: https://doi.org/10.1182/blood-2013-08-519124
- Primary Citation of Related Structures:
4KI5 - PubMed Abstract:
The factor VIII C2 domain is a highly immunogenic domain, whereby inhibitory antibodies develop following factor VIII replacement therapy for congenital hemophilia A patients. Inhibitory antibodies also arise spontaneously in cases of acquired hemophilia A. The structural basis for molecular recognition by 2 classes of anti-C2 inhibitory antibodies that bind to factor VIII simultaneously was investigated by x-ray crystallography. The C2 domain/3E6 FAB/G99 FAB ternary complex illustrates that each antibody recognizes epitopes on opposing faces of the factor VIII C2 domain. The 3E6 epitope forms direct contacts to the C2 domain at 2 loops consisting of Glu2181-Ala2188 and Thr2202-Arg2215, whereas the G99 epitope centers on Lys2227 and also makes direct contacts with loops Gln2222-Trp2229, Leu2261-Ser2263, His2269-Val2282, and Arg2307-Gln2311. Each binding interface is highly electrostatic, with positive charge present on both C2 epitopes and complementary negative charge on each antibody. A new model of membrane association is also presented, where the 3E6 epitope faces the negatively charged membrane surface and Arg2320 is poised at the center of the binding interface. These results illustrate the potential complexities of the polyclonal anti-factor VIII immune response and further define the "classical" and "nonclassical" types of antibody inhibitors against the factor VIII C2 domain.
- Western Washington University, Department of Chemistry, Bellingham, WA; and.
Organizational Affiliation: