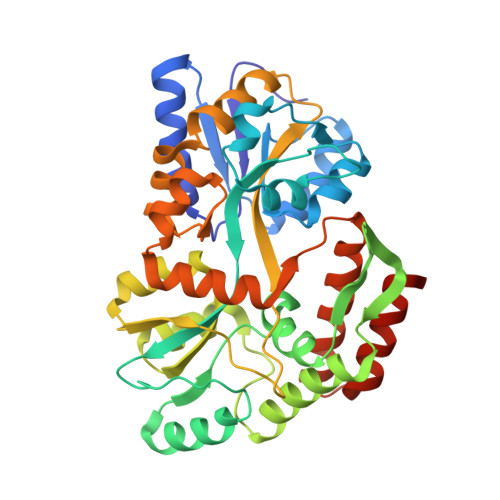
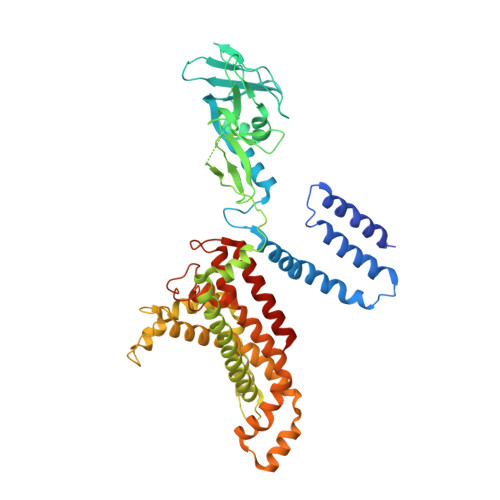
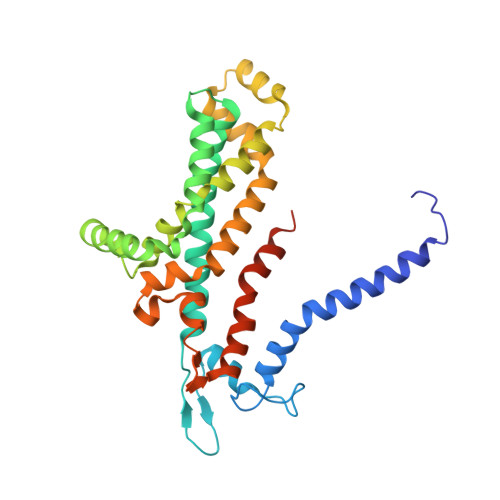
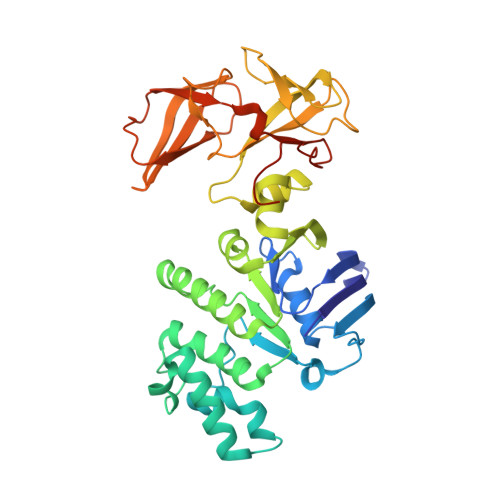
Structural basis for substrate specificity in the Escherichia coli maltose transport system.
Oldham, M.L., Chen, S., Chen, J.(2013) Proc Natl Acad Sci U S A 110: 18132-18137
- PubMed: 24145421
- DOI: https://doi.org/10.1073/pnas.1311407110
- Primary Citation of Related Structures:
4KHZ, 4KI0 - PubMed Abstract:
ATP-binding cassette (ABC) transporters are molecular pumps that harness the chemical energy of ATP hydrolysis to translocate solutes across the membrane. The substrates transported by different ABC transporters are diverse, ranging from small ions to large proteins. Although crystal structures of several ABC transporters are available, a structural basis for substrate recognition is still lacking. For the Escherichia coli maltose transport system, the selectivity of sugar binding to maltose-binding protein (MBP), the periplasmic binding protein, does not fully account for the selectivity of sugar transport. To obtain a molecular understanding of this observation, we determined the crystal structures of the transporter complex MBP-MalFGK2 bound with large malto-oligosaccharide in two different conformational states. In the pretranslocation structure, we found that the transmembrane subunit MalG forms two hydrogen bonds with malto-oligosaccharide at the reducing end. In the outward-facing conformation, the transmembrane subunit MalF binds three glucosyl units from the nonreducing end of the sugar. These structural features explain why modified malto-oligosaccharides are not transported by MalFGK2 despite their high binding affinity to MBP. They also show that in the transport cycle, substrate is channeled from MBP into the transmembrane pathway with a polarity such that both MBP and MalFGK2 contribute to the overall substrate selectivity of the system.
- Howard Hughes Medical Institute and Department of Biological Sciences, Purdue University, West Lafayette, IN 47907.
Organizational Affiliation: